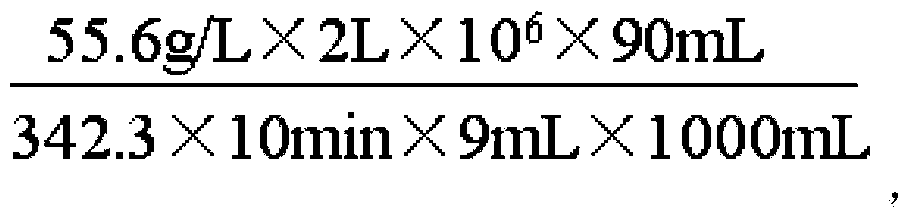Genetic engineering bacteria used for high efficient production of mycose
A technology of genetically engineered bacteria and trehalose, applied in the fields of genetic engineering and enzyme catalysis, can solve the problems of high enzyme cost, long conversion time, and insufficient enzyme activity, and achieve the effects of low enzyme cost, short conversion time, and simple process operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1. Knockout of treA and malQ genes on the chromosome genome of Escherichia coli BL21 (DE3)
[0034] 1.1. Experimental route
[0035] In the present invention, the host bacteria Escherichia coli BL21 (DE3) is genetically engineered, and the trehalase gene (treA) and malt glucosylase gene (malQ) located on the chromosome genome are knocked out. The gene is knocked out step by step, and the knockout of the two genes is carried out according to the following method:
[0036] The first step: the construction of the resistance gene targeting fragment;
[0037] The second step: replace the treA gene or malQ gene with the resistance gene targeting fragment;
[0038] Step 3: Eliminate the resistance gene.
[0039] 1.2. Construction of treA resistance gene targeting fragment
[0040] Synthetic primers P1 and P2:
[0041] P1: 5'-AACGCCGCTGGCTGGCTAAT-3' (SEQ ID NO: 1);
[0042] P2: 5'-TATCTTCGGCAACCACGTAT-3' (SEQ ID NO: 2).
[0043] Prepare a PCR reaction solution c...
Embodiment 2
[0058] Example 2 , Construction of (Escherichia coli) BL21(DE3)MTSaseMTHase
[0059] The whole gene synthesis is derived from the maltooligosaccharide-based trehalose synthase gene MTSase and the maltooligosaccharide-based trehalose hydrolase gene MTHase from Arthrobacter, Sulfolobus, Brevibacterium, Mycobacterium, subcloned into expression plasmid vectors (such as pTrc99a and pET24a, etc.), the plasmid vector containing the polynucleotide sequence encoding MTSase and the plasmid vector containing the polynucleotide sequence encoding MTHase were obtained respectively. Then use two kinds of suitable restriction endonucleases to digest the plasmid vector mixture containing the MTSase polynucleotide sequence encoding various sources, and use the same restriction enzyme to digest the plasmid vector containing the MTHase polynucleotide sequence encoding various sources Plasmid carrier mixture, obtain the fragment containing the MTSase polynucleotide sequence and the fragmen...
Embodiment 3
[0061] Example 3 , Using CCTCC NO: M2013237 to produce trehalose
[0062] Insert the CCTCC NO: M2013237 strain into a shaker flask containing 50mL of seed medium at a volume ratio of 1‰, and culture it at 37°C and 200 rpm for 12 hours, when OD600=1~2 stop the culture, put it into a shake flask with 1L of fermentation medium, culture it at 37°C and 200 rpm for 3 hours, lower the culture temperature to 28°C, and add isopropyl-β- D-thiogalactopyranoside, continue to cultivate for 20 hours, and finish the fermentation. The bacterial cells in the fermentation broth were collected by centrifugation, and the wet weight was 30g / L. 2 times the deionized water was added according to the mass, and after mixing evenly, the cells were broken by an ultrasonic cell breaker to obtain 90ml cell breakage solution.
[0063] Add cornstarch milk (starch mass concentration: 31%) at pH 5.3 to 5.6, add high-temperature amylase (Liquozyme supra, purchased from Novozymes) at an amount of 250 mL / ton ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com