Method, primer pair, target probe, internal standard probe and kit for detecting Cronobacter sakazakii
A technology of internal standard probes and detection methods, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of long detection time, time-consuming, heavy workload, etc., and achieve short detection time and high results The effect of simple judgment and reduced inspection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Primer, Internal Standard Sequence, and Probe Synthesis
[0044] Synthesize primers, internal standard sequences (synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd.) and probes (Shanghai Biotechnology Co., Ltd.) capable of PCR amplifying Cronobacter sakazakii specific gene sequence (CS44) company), the sequence is as follows:
[0045] SEN2-L (primer): 5'-aatgcacaagcgcgatttc-3' (SEQ ID NO: 1)
[0046] SEN2-R (primer): 5'-tgcaaaaggcgctgtgataa-3' (SEQ ID NO: 2)
[0047] Probe-1 (target probe): 5'-FAM-tgctcgcccaatctcaacgcc-BHQ1-3' (SEQ ID NO:3)
[0048] Probe-2 (internal standard probe): 5’-HEX-ccttagtaaaaccggtacctc-BHQ1-3’SEQ ID NO:4)
[0049] Internal standard template:
[0050] 5'-aatgcacaagcgcgatttccacgccttagtaaaaccggtacctctacaattatcacagcgccttttgca-3' (SEQ ID NO: 5)
Embodiment 2
[0052] Detection of Cronobacter sakazakii
[0053] 1. Suggestions and reaction parameters for PCR detection reaction system
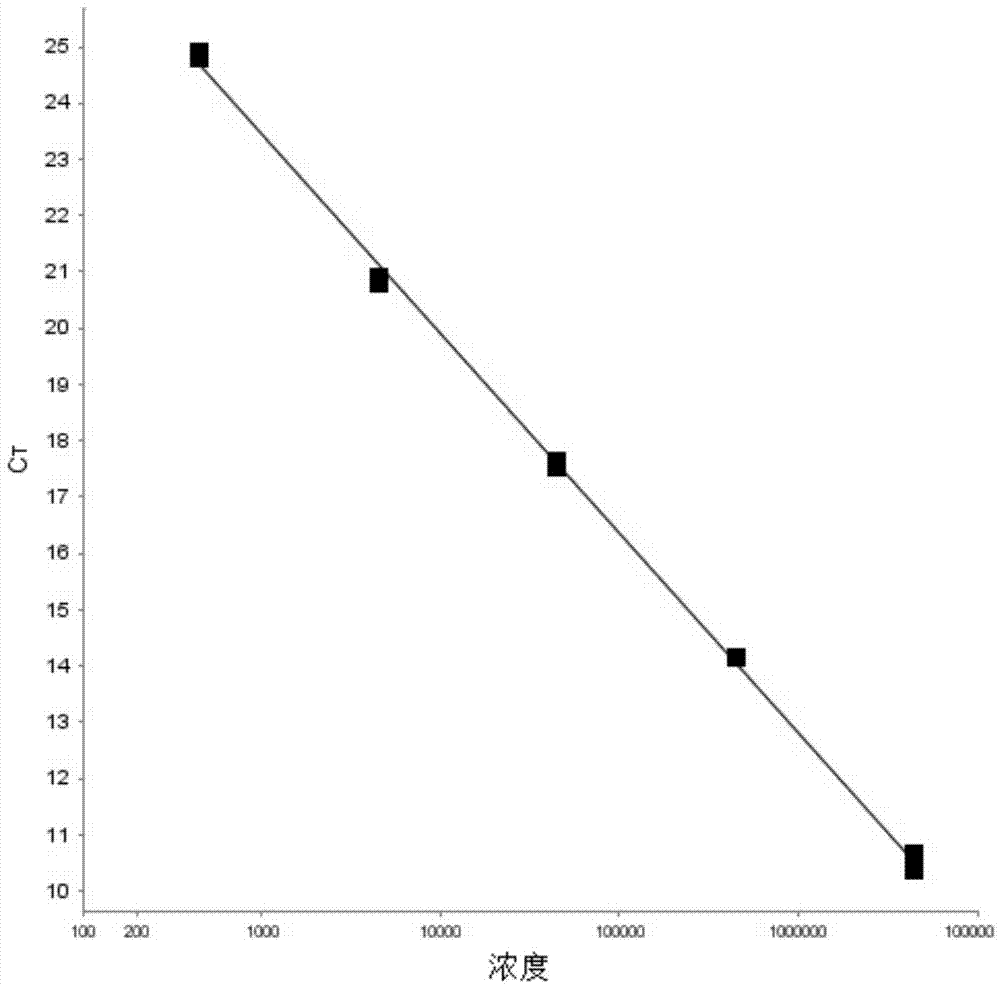
[0054] Using the primers and probes as described in Example 1, the genomic DNA of the standard strain of Cronobacter sakazakii ATCC29544 (purchased from ATCC) was used as a template, and the PCR reaction reagent was provided by Promega Probe qPCR Master Mix (Catalog No. A6102), by adjusting and optimizing the concentration of primers, target probes and internal standard probes in the reaction system, the PCR reaction system can achieve the best reaction system. The concentration of the primers is 400 nM, and the concentration of the genomic DNA of the sample to be tested is 20 ng / μL.
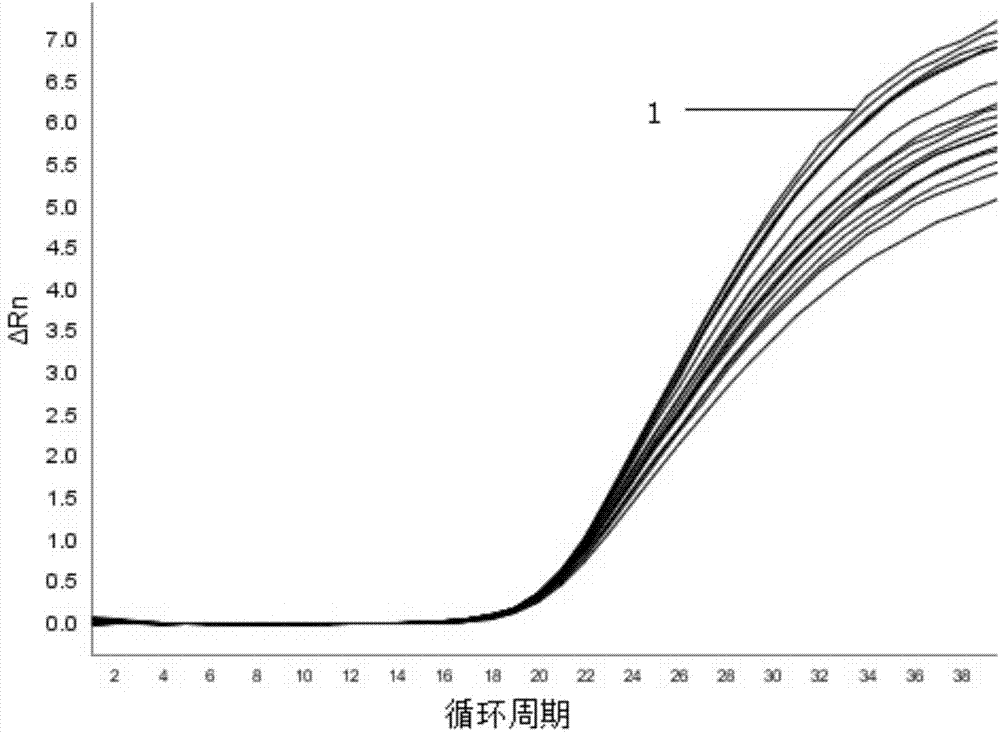
[0055] Add target probes and internal standard probes with an original concentration of 10 μM to different PCR reaction tubes, and serially dilute the concentrations of target probes and internal standard probes from 100 nM to 500 nM (100 nM, 150 nM, 200 nM, 250 nM, 30...
Embodiment 3
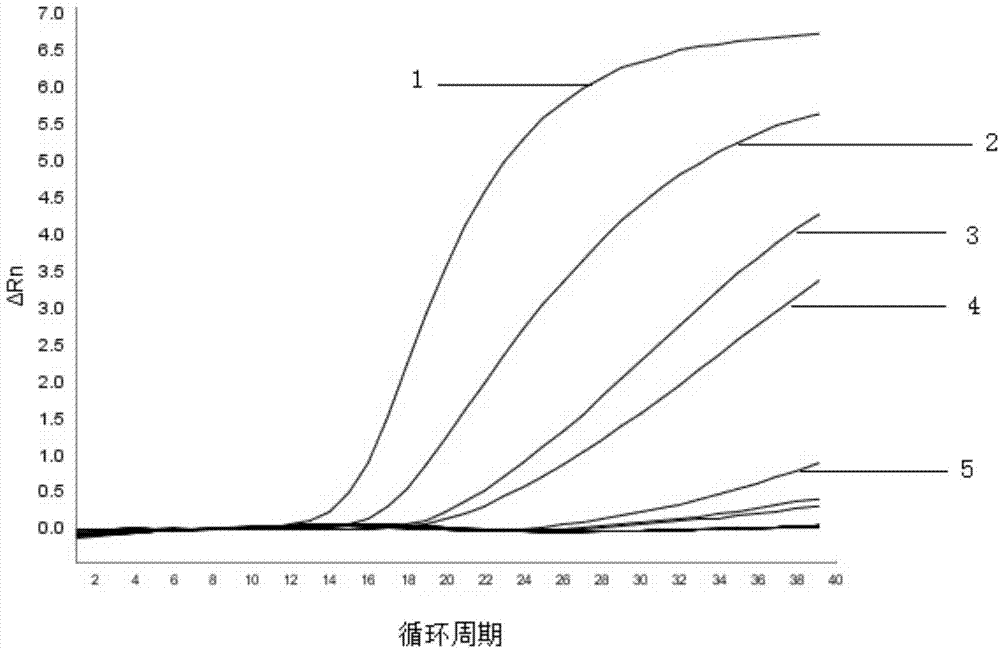
[0071] The 30 food samples (including powdered milk and vegetables) were purchased from farmers' markets and large supermarkets in Shanghai. Add 50g of each of the 30 samples to 450mL buffer peptone water (buffer peptone water, BPW, pH8.0) enrichment medium (ingredients include: peptone 10.0g, sodium chloride 5.0g, disodium hydrogen phosphate 9.0g , Potassium dihydrogen phosphate 1.5g, distilled water 1000mL, pH7.2±0.2; the preparation method is: add each ingredient into distilled water, stir well, let stand for about 10min, boil to dissolve, adjust pH, autoclave at 121℃, 15min) After enrichment at 37°C for 18 hours, take 1 mL of each sample and put it into a 1.5 mL centrifuge tube, and extract genomic DNA by boiling method, centrifuge at 12,000 rpm for 5 min, and take 2.5 μL of supernatant as a PCR template. Sterile water was used as a negative control for fluorescent quantitative PCR detection.
[0072] The reaction system for PCR detection is as shown in Example 1; the rea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com