High-salt-tolerance Zygosaccharomyces rouxii A
A technology of Zygosaccharomyces rouckeri and Zygosaccharomyces, which is applied in the field of microorganisms, can solve the problems that yeast cannot adapt to the high-salt environment of soy sauce and cannot function well, and achieve shortened fermentation cycle, good growth and high salt tolerance Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: 26S rRNA identification experiment of Lu's conjugation yeast A of the present invention
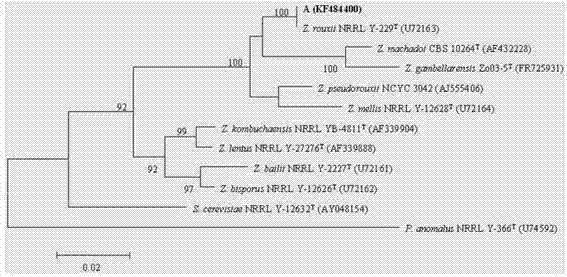
[0025] The extraction and purification of yeast genomic DNA used the method of Sambrook et al. (1989). Using extracted and tested qualified chromosomal DNA as a template, NL 1 (5'-GCATATCAATAAGCGGAGGAAAAG-3') and NL4 (5'-GGTCCGTGTTTCAAGACGG-3') were selected as primers. The PCR amplification of 26s rRNA gene sequence was performed according to Solieri L. et al. (2007) Method operation, gene sequence determination was completed by Shanghai Yingjun Company. Using Clustalx1.8, MEGA 5.0 software (Tamura et al. 2011), according to the results of the 26S rRNA gene sequence similarity analysis, the 26S rRNA gene sequences of related strains were selected, and neigbor-joining (neighbor-joining method, NJ) ( Saitou and Nei 1987) algorithm to get the corresponding phylogenetic tree ( figure 2 ). The constructed phylogenetic tree shows that strain A is in Zygosaccharomyces On a br...
Embodiment 2
[0026] Example 2: Characteristic analysis experiment of fatty acid composition of whole cell of Lu's conjugative yeast A according to the present invention
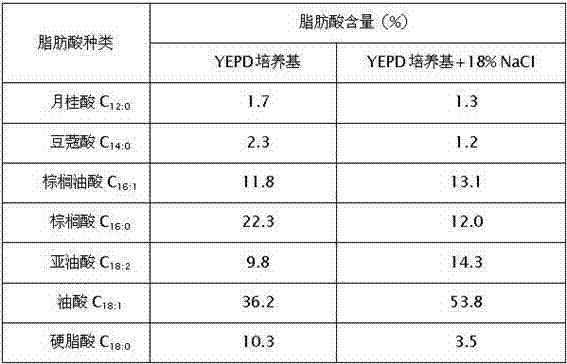
[0027] Yeast relies on the saturation of its cell membrane fatty acids to adapt to the unfavorable factors such as alcohol, salt, and high temperature in the surrounding environment. In order to analyze the fatty acid composition of strain A, strain A was first inoculated into YEPD broth, cultured at 28°C to the late logarithmic growth phase, and appropriate bacteria were collected. According to "the protocol of the Sherlock Microbial Identification System (MIDI)" (version) 6.0.) The method described extracts the methylated esters of bacterial fatty acids, and finally uses Agilent 6890N GC gas chromatography to analyze the fatty acid composition through the MIDI Sherlock YEAST6 database (Sasser, 1990). The fatty acid composition of the whole cell of strain A of the present invention is shown in Table 1. Strain A whole cell ...
Embodiment 3
[0030] Example 3: Salt tolerance experiment of Lu's conjugative yeast A of the present invention
[0031] Prepare medium (1% yeast powder, 5% glucose, 2% peptone, KH 2 PO 4 0.1%, KCl 0.04%, CaCl 2 0.11%, (NH 4 ) 2 SO 4 0.5%, MnSO 4 0.25 mg%, FeCl 3 0.25 mg%), add NaCl, set the salt concentration to 16%, 18%, 20%, 21%, 22%, 23%, 24%, 25%, 26%. After moist heat sterilization, it is connected to the present invention Strain A was cultured at 28°C at 200 rpm to observe the time when the medium became turbid. The results are shown in Table 2. The strain A of the present invention grows well in the presence of 20% sodium chloride, can maintain a certain amount of growth in the presence of 24% sodium chloride, and can still grow in the presence of 26% sodium chloride. However, the existing Lu's Zygogium generally grows well in the presence of less than 16% sodium chloride, and can hardly grow in the presence of 20% sodium chloride.
[0032] Table 2 Salt tolerance test of strain A of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com