DNA polymerase coding DNA, enzyme coded thereby, and application and preparation method of DNA polymerase
A polymerase and coding technology, applied in the field of genetic engineering, can solve the problems of high GC template can not be well amplified, the amplification error rate is increased, time-consuming and laborious samples, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Cloning and expression of embodiment 1.Pyrococcus yayanosii DNA polymerase gene
[0032] The strains were purchased from the Japanese Culture Collection of Microorganisms (preservation number: JCM16557), centrifuged at 12000rpm for 5min, and the supernatant was discarded; the cell pellets refer to Genomic DNA Purification kit (Promega Company) was used to extract genomic DNA. The brief steps are as follows: Cell pellets were resuspended in 600 μL Nuclei Lysis Solution (nuclei lysis solution), and cells were lysed in a water bath at 80°C for 5 minutes; after cooling to room temperature, 200 μL Protein Precipitation Solution (protein precipitation solution) was added. solution), shake and mix, ice bath for 5min; centrifuge at 12000rpm for 5min; pipette the supernatant into another clean centrifuge tube, add 600μL isopropanol, mix well, and ice-bath for 10min; centrifuge at 12000rpm for 5min; wash the nucleic acid pellet with 700μL70% ethanol Twice, after centrifugation a...
Embodiment 2
[0035] The purification of embodiment 2.DNA polymerase Pya
[0036] Inoculate the bacteria that can correctly express the DNA polymerase Pya into a 500mL Erlenmeyer flask containing 200mL LB medium at a ratio of 1:100, add kanamycin and 34mg / L chloramphenicol at a final concentration of 30mg / L, and store at 37°C Shake culture at 200rpm until OD600=0.4, add IPTG with a final concentration of 0.1mM to induce the expression of the target protein, and induce at 30°C for 4h; collect the induced bacterial liquid, centrifuge at 12000rpm for 2min, discard the supernatant, and use 20mL buffer A (20mM Tris- HCl (pH7.8), 0.1M KCl, 50mM NaCl) plus 30mM imidazole were resuspended, and stored in a -80°C refrigerator for 12 hours; lysed at 37°C, ultrasonically disrupted, treated in a 70°C water bath for 30min, and centrifuged at 12,000rpm for 10min; The supernatant was passed through nickel affinity medium (IDA, National Biochemical Engineering Technology Research Center), and eluted with bu...
Embodiment 3
[0037] Example 3. Application of DNA polymerase Pya in nucleic acid amplification
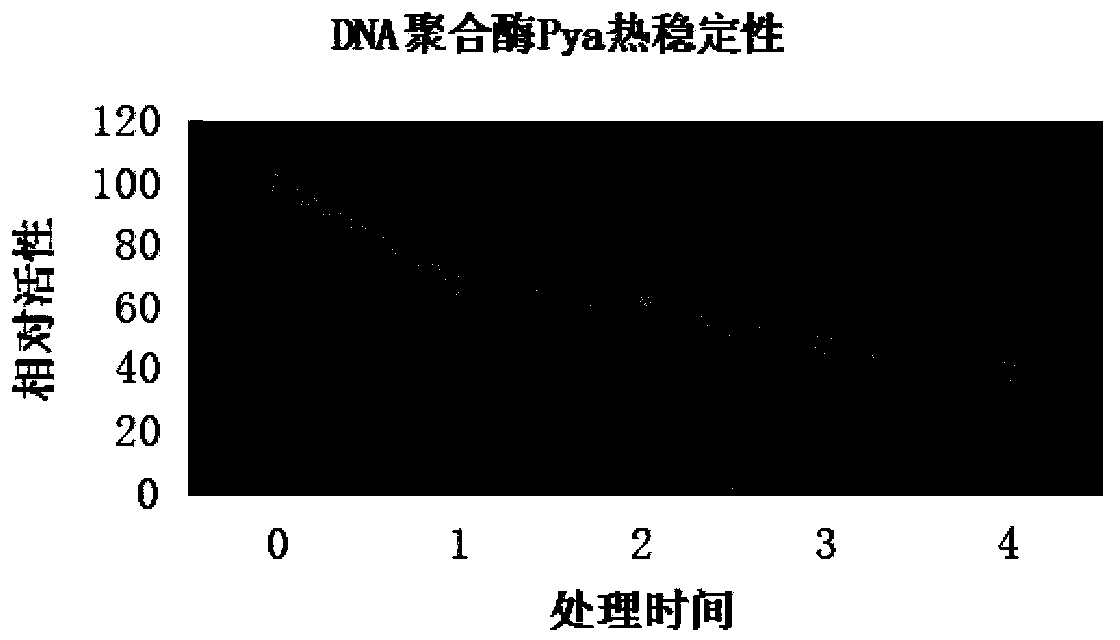
[0038] The activity of the enzyme obtained in Example 2 was determined by the amount of nucleotides incorporated at 72°C for 30 minutes, diluted to 2.5u / μL, and its 3'-5' exonuclease activity was tested by the fluorescent probe method. As a result, Pya has a strong The correcting activity of , indicating that its amplification fidelity performance is better. In order to test the thermal stability of DNA polymerase Pya, the enzyme was incubated at 99°C, and the enzymes treated for 0, 1, 2, 3, and 4 hours were used to test the DNA polymerase activity. The results showed that DNA polymerase Pya had extremely high thermal stability. Its 99°C half-life is 3h ( figure 2 ).
[0039] Test the reaction conditions of DNA polymerase, use the pUC19 plasmid as a template, design primers to amplify about 2.7kb fragment by PCR to determine the optimal pH and K of the reaction + , NH4 + , Mg 2+ ion conce...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com