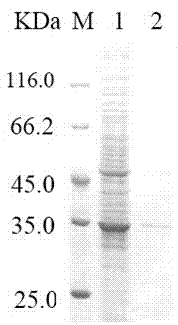
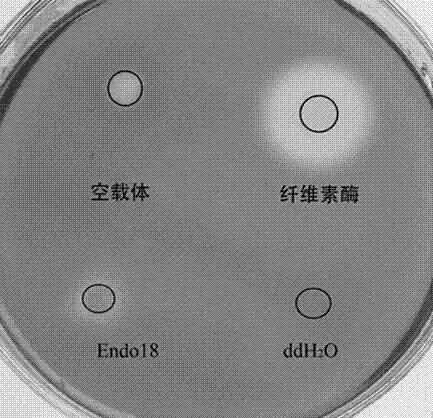
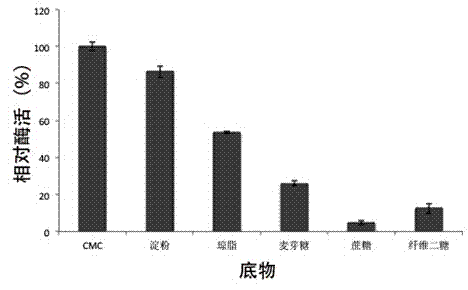
Alkaline cellulase, and DNA (deoxyribonucleic acid) sequence and application thereof
A cellulase and cellulose degradation technology, applied in the field of peptides, can solve the problems of difficulty in glucose and the limitation of cellulose regeneration and utilization, and achieve the effects of good alkali-resistant cellulase activity and broad industrial application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] The acquisition of metagenomic DNA in the soil sample of embodiment 1
[0044] Papermaking wastewater sediment samples were collected from the outfall of a paper mill in Xuchang City, Henan Province.
[0045] Take two 50 mL centrifuge tubes, weigh papermaking wastewater sediment samples, 6 g in each tube, add DNA extraction buffer 13.5 mL (containing 100 mM EDTA, 100 mM sodium phosphate, 1.5 M NaCl, 1% CTAB and 100 mM Tris-Hcl, pH 8.0), vortexed to mix, placed on a shaker at 220 rpm, activated at 37°C for 30 min, then added 1.5 mL of 20% SDS to each tube to make the final concentration 2% (w / v) . Then, bathe in a water bath at 65°C for 2 h, and gently invert up and down several times every 15 min to mix well. Centrifuge at 6,000 g at room temperature for 10 min. After collecting the supernatant, transfer it to a clean centrifuge tube, add an equal volume of chloroform:isoamyl alcohol (24:1, V / V), and gently invert up and down to mix. Then centrifuge at 11,000 g at ...
Embodiment 2
[0046] Example 2 Construction of the Metagenome Library of Papermaking Wastewater Sediments and Acquisition of DNA Sequences
[0047] 1. Construction of metagenomic library of papermaking wastewater sediment
[0048] use Eco The RI enzyme partially digested the metagenomic DNA of the papermaking wastewater sediment extracted in Example 1, and electrophoresis recovered a 2.5-7.5 kb digested fragment. Combine the recovered DNA fragments with the Eco RI digested and dephosphorylated pUC118 vector (Takara company) ligation, electroporation transformation E. coli DH5α competent cells were spread on LB solid plates containing 100 μg / mL ampicillin (Amp), 40 μg / mL X-gal and 0.5 mM IPTG, thereby constructing a library with a capacity of 10 7 Metagene library with multiple transformants and good diversity.
[0049] 2. Screening and sequence analysis of target genes
[0050] Randomly pick the clones on the plate and transfer them in parallel to CMC plates containing 100 μg / mL A...
Embodiment 3
[0052] Example 3 Obtaining of recombinant protein expressed by DNA sequence
[0053] 1. Inoculate the positive clones obtained in Example 2 into 5 mL LB medium (100 μg / mL Amp), culture on a shaker at 37 °C and 220 rpm for 12 h, and extract the plasmid.
[0054] 2. Design primers P1 and P2. Both ends of the primers are respectively introduced into the Saccharomyces cerevisiae expression vector pyes2 Eco R I and xho I Restriction site.
[0055] The nucleotide sequence of primer P1 (upstream primer) is shown in SEQ ID NO:3:
[0056] The nucleotide sequence of primer P2 (downstream primer) is shown in SEQ ID NO:4:
[0057] 3. Using the extracted plasmid as a template, PCR amplification was performed using primers P1 and P2.
[0058] The total volume of the PCR system is 50 μL, including Prime STAR TM Max Premix 25 μL (2.5 U), two primers each 2 μL (10 μM), sterilized distilled water 20 μL and template DNA 1 μL (1 ng).
[0059] PCR amplification conditions: 98°C pre-denat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com