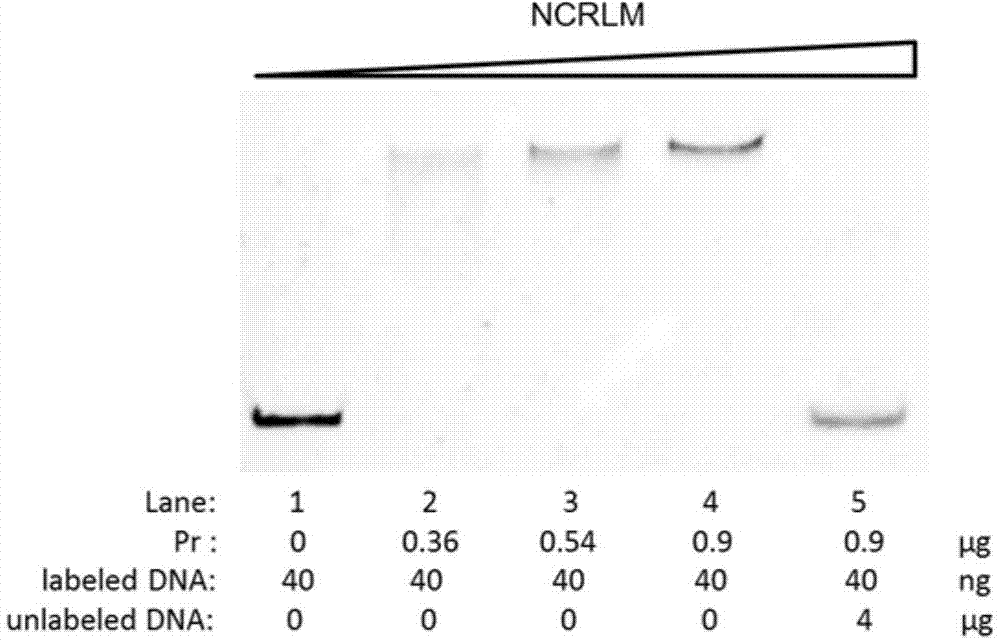
Nosiheptide biosynthetic regulatory protein NosP specific binding sequence and related application thereof
A technology of biosynthesis and combination of sequences, which is applied in the fields of application, biochemical equipment and methods, plant gene improvement, etc., can solve the problems such as the unknown influence of DNA sequence nosipeptide biosynthesis titer, so as to improve fermentation titer and reduce production cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Using the S. actuosus ATCC 25421 genome as a template, the 5'-end truncated nosP gene lacking 207 bases was amplified by PCR, and restriction endonuclease sites were inserted in the upstream and downstream of the gene sequence, using the technology in the art The familiar enzyme digestion-ligation method is used to construct recombinant expression plasmids containing different selection markers: Scheme 1: With the help of restriction sites NdeI / HindIII and pET22b(+), insert a histidine tag at the C-terminus of NosP to obtain a recombinant plasmid Is pET22b-his-c; Scheme 2: With the help of restriction sites NdeI and HindIII, insert a histidine tag at the N-terminus of NosP to obtain the recombinant plasmid pET28a-his-n; Scheme 2: With the help of restriction sites BamHI and XhoⅠ was inserted into the GST tag at the N-end of NosP, and the recombinant plasmid was obtained as pGEX4T-gst-n. After sequencing and verification, the above recombinant plasmid was transferred into ...
Embodiment 2
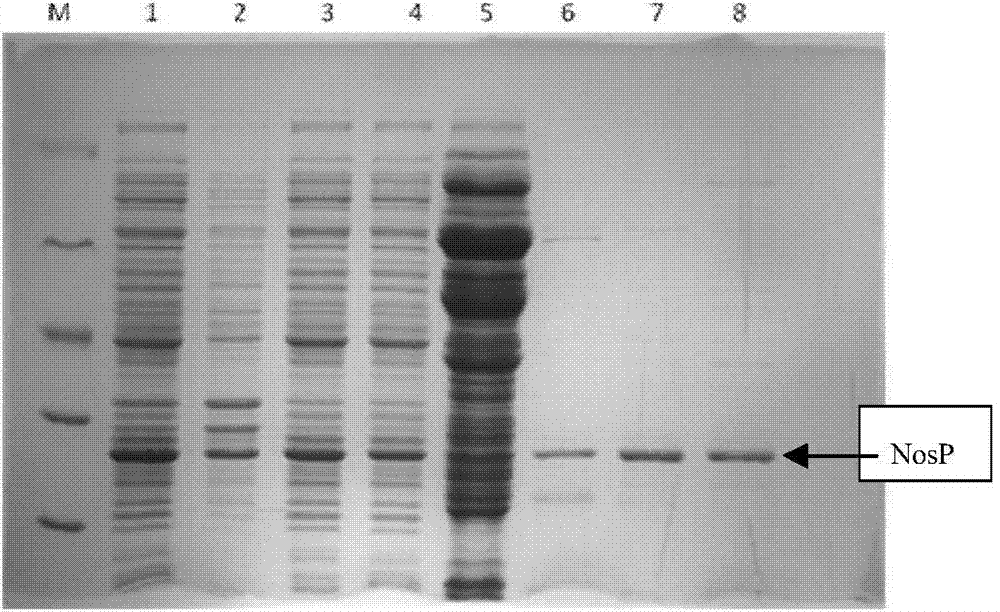
[0020] The three genetic recombinant E.coli obtained in Example 1 were cultured in 50mL LB medium (containing 100μg / mL ampicillin or 50μg / mL kanamycin), shaking overnight at 37°C and 220rpm to obtain the corresponding Seed liquid. Transfer the seed solution to LB medium (containing 100μg / mL ampicillin or 50μg / mL kanamycin) according to the inoculum amount of 5%, and shake culture to the OD of the medium at 37℃ 600 When =0.8±0.1, add the inducer IPTG with a final concentration of 0.001-2mM, induce culture at 25°C for 20 hours to obtain the engineered strain E.coli fermentation broth. The fermentation cells were collected by centrifugation at 4°C and 4500 rpm for 15 min. The cells were washed twice with 40 mM Tris-HCl buffer (pH 8.0), and the cells were collected for SDS-PAGE analysis. The results showed that the three engineering strains of pHC, pHN and pHG all obtained the best soluble expression under the condition of 0.2mM IPTG. The content of NosP accounted for 8.5%, 7.8% an...
Embodiment 3
[0022] The three genetic recombinant E.Coli obtained in Example 1 were cultured in 50mL LB medium (containing 100μg / mL ampicillin or 50μg / mL kanamycin), shaking overnight at 37°C and 220rpm to obtain the corresponding Seed liquid. Transfer the seed solution to LB medium (containing 100μg / mL ampicillin or 50μg / mL kanamycin) according to the inoculum amount of 5%, and shake culture to the OD of the medium at 37℃ 600 When =0.8±0.1, add the inducer IPTG with a final concentration of 0.2mM, and induce culture at 10~37℃ for 20 hours to obtain the engineered strain E.coli fermentation broth. The fermentation cells were collected by centrifugation at 4°C and 4500 rpm for 15 min. The cells were washed twice with 40 mM Tris-HCl buffer (pH 8.0), and the cells were collected for SDS-PAGE analysis. The results showed that the soluble expression of NosP in the three engineering strains of pHC, pHN and pHG reached the maximum at 16°C, accounting for 9.3%, 8.4% and 8.1% of the total protein, r...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com