Rice genome-wide breeding chip and its application
A whole-genome and rice technology, applied in the fields of genomics, molecular biology, bioinformatics and molecular plant breeding, can solve the problems of large-scale commercial breeding, cumbersome operation process, low throughput, etc., and achieve sequencing cost reduction, simple data analysis, and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1Rice60K rice whole genome breeding chip preparation method
[0044] The present invention utilizes the Illumina sequencing technology to test 211 rice varieties, see Table 1. Whole-genome sequencing was performed to obtain 2.5-fold genome coverage per sample. At the same time, the present invention also downloaded the sequencing data of 520 rice landraces from the public database (http: / / www.ebi.ac.uk / ena / , accession numbers ERP000106, ERP000235, ERP000236), each sample 1 times the genome Coverage (Huang et al., Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010, 42:961-967). A total of 731 rice cultivar genome sequencing data. Follow the steps below to identify and screen SNPs.
[0045] Table 1 Names of 211 rice varieties
[0046]
[0047]
[0048]
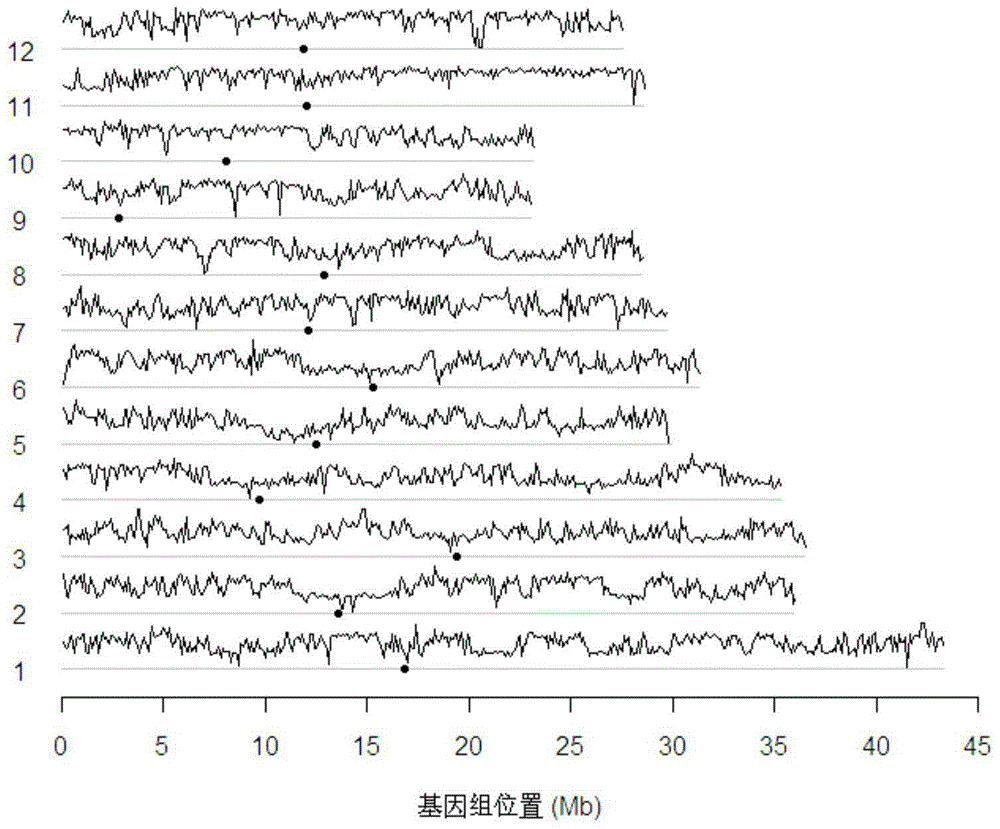
[0049] 1. 3,791,687 high-quality SNP sites were identified from the sequencing data of 731 rice varieties. High-quality SNP sites meet the following conditio...
Embodiment 2
[0057] Example 2 Application of Rice60K rice genome-wide breeding chip in detecting rice DNA samples
[0058] Rice genomic DNA extraction: Genomic DNA is extracted from rice seeds, leaves and other tissues according to the detection requirements. Among them, it is recommended to use Promega Plant Genome Extraction Kit (Wizard Magnetic 96DNAPlant System Kit, Cat. No. FF3760 or FF3761, Beijing Promega Biotechnology Co., Ltd.) for DNA extraction of young rice leaves, and it is recommended for DNA extraction of rice seed endosperm SurePlant Genome Extraction Kit (SurePlant DNA Kit, Cat. No. CW0555, Beijing Kangwei Century Biotechnology Co., Ltd.) was used for extraction.
[0059] DNA sample quality detection: use agarose gel electrophoresis detection with a mass fraction of 1-1.5% (W / W), and use a gel imaging system (Gel Doc XR System, Bio-Rad, USA) to systematically judge the electrophoresis results to ensure that the genome DNA integrity is good, and the length of the genomic D...
Embodiment 3
[0062] Example 3 Application of Rice60K Rice Whole Genome Breeding Chip in Genomic Fingerprint Analysis of Rice Germplasm Resources
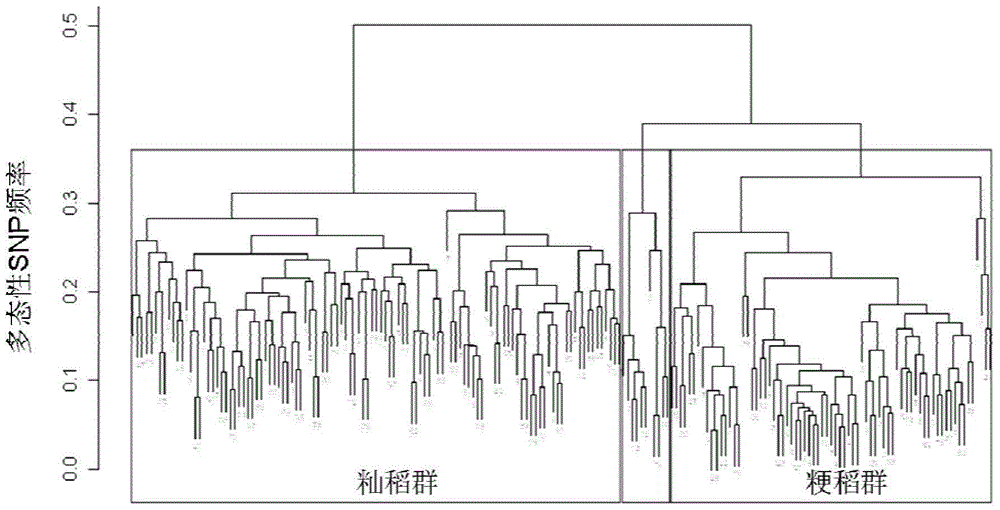
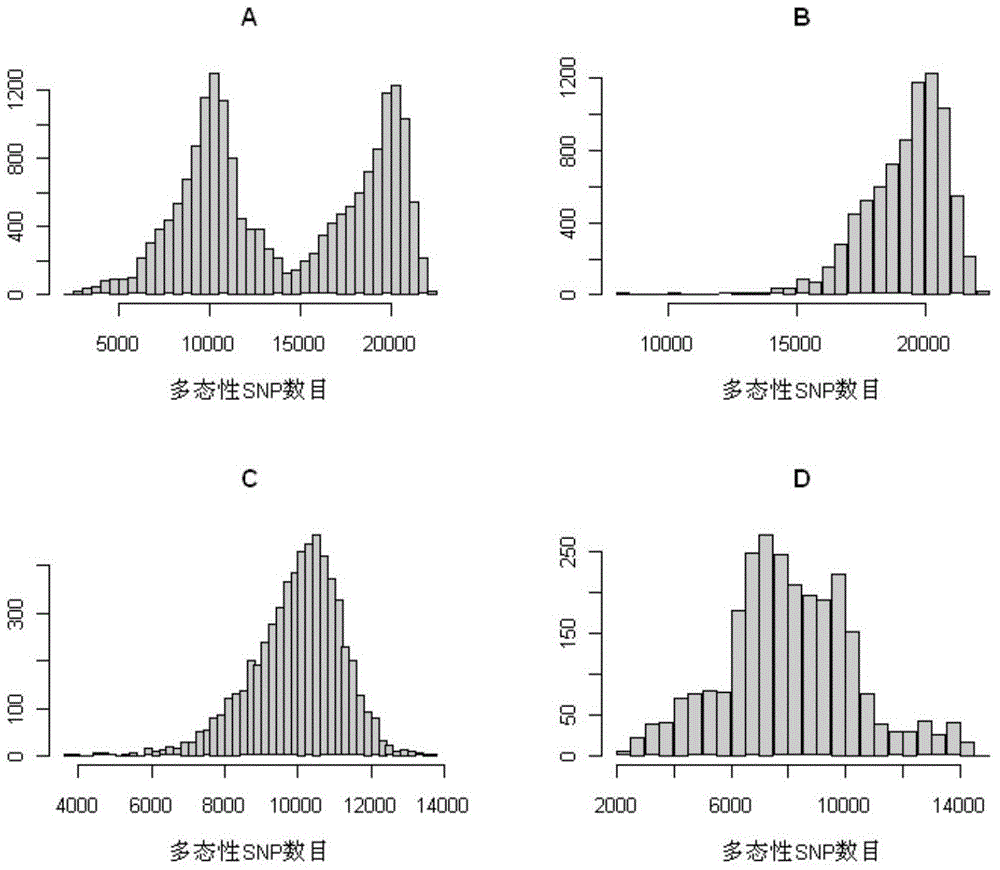
[0063] The invention uses the Rice60K breeding chip to pair 270 varieties of micro-core rice germplasm resources, numbered WCR001-270, from China Seed Group Co., Ltd. Detection was carried out with reference to the method of Example 2, and 195 representative homozygous inbred varieties were selected for analysis according to the detection results. According to the genotype detected by the Rice60K breeding chip, the cluster analysis of these 195 varieties can be divided into three categories: indica rice, japonica rice and intermediate types (see figure 2 ), which is in good agreement with the classification according to the phenotype. The average numbers of polymorphic SNPs between any two varieties, between indica and japonica, between indica and indica, and between japonica and japonica were about 14,000, 19,000, 10,000, and 8,000, respectiv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com