A kind of alginate lyase sha-5 gene and its prokaryotic expression vector
An alginate lyase, SHA-5 technology, applied in genetic engineering, plant genetic improvement, introduction of foreign genetic material using vectors, etc., can solve the problem of high cost, difficult to meet application requirements, and wild-type alginate decomposing bacteria to produce enzymes. It is easy to operate, has broad substrate specificity, and is convenient for industrial production.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1: Marinicatena alginatilytica Preparation and Detection of SH-52 Genomic DNA
[0037] used in the present invention Marinicatena alginatilytica SH-52 is a strain screened by our laboratory. The preparation of SH-52 genomic DNA adopts the extraction method of common bacterial genome. The specific content is as follows: Take 2 mL of overnight culture liquid and centrifuge at 4000 rpm for 2 min at 4 ° C. Discard the supernatant and collect the bacteria. Add 100ul Solution I suspension bacteria, 30ul 10% SDS and 1ul 20mg / ml proteinase K, mix well, and incubate at 37°C for 1 hour; add 100ul 15mol / L NaCl, mix well; add 20ul CTAB / NaCl solution (CTAB 10%, NaCl 0.7mol / L), mix well, 65℃, 10 minutes; add an equal volume of phenol / chloroform / isoamyl alcohol (25:24:1) to mix, centrifuge at 12000rpm for 5 minutes; take the supernatant, add 2 times the volume of Water and ethanol, 0.1 times the volume of 3mol / L NaOAC, placed at -20°C for 30 minutes; centrifuged at 12,000 ...
Embodiment 2
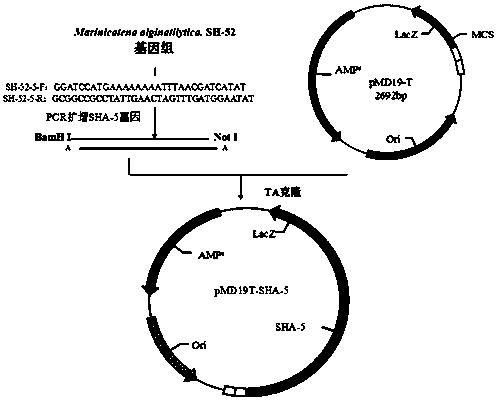
[0038] Example 2: Alginate Lyase SHA-5 Gene amplification and TA cloning
[0039] alginate lyase SHA-5 gene amplification and cloning figure 2 As shown, first find out from the whole genome sequencing results SHA-5 The full-length gene sequence, and design a pair of specific primers, the sequence is as follows:
[0040] SHA-5-F: GGATCC ATGAAAAAAAATTTAACGATCATAT
[0041] SHA-5-R: GCGGCCGC CTATTGAACTAGTTTGATGGAATAT
[0042] The 5' end primer has the GGATCC characteristic sequence, and thus forms the BamH I restriction site; the 3' end adds the GCGGCC characteristic sequence, forming the Not I restriction site.
[0043] Add 10 ng of Marinicatena alginatilytica SH-52 genomic DNA is used as a template, and 50ng of specific primers SHA-5-F and SHA-5-R, 2.5μl of dNTP (10mM), 2.5μl of Pfu reaction buffer and 0.5μl of pfu (5U / ul) are added at the same time Polymerase (Beijing Quanshijin Biotechnology Co., Ltd.), add double distilled water to make the final volume 25 μl. H...
Embodiment 3
[0044] Example 3: Prokaryotic expression vector pGEX-4T-1- SHA-5 build
[0045] pGEX-4T-1- SHA-5 A build strategy such as Image 6 As shown, the purified prokaryotic expression vectors pGEX-4T-1 (purchased from GE Healthcare) and pMD19T- SHA-5 , separated the cut vector and insert fragments by agarose gel electrophoresis, and recovered the vector fragments pGEX-4T-1 (4.9kb) and pMD19T- SHA-5 produced by cutting SHA-5 The DNA fragment of the gene (about 1.7kb), and then use the ligase kit of TaKaRa to connect the pGEX-4T-1 vector fragment and SHA-5 The DNA fragment of the gene produces the prokaryotic expression vector pGEX-4T-1- SHA-5 . Use the ligation reaction mixture to transform high-efficiency E. coli competent cells Trans1-T1 (Beijing Quanshijin Biotechnology Co., Ltd.), and spread the transformed E. coli on a plate added with ampicillin (Amp, 100mg / L). Cultivate overnight at 37°C, screen Amp-resistant recombinant colonies, and extract plasmids from Amp-resistant ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com