SSR molecular marker method for identifying foxtail millet variety and application
A technology of molecular markers and millet, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of high cost, long cycle, and seasonal restrictions, and achieve better results and save time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1: the establishment of method
[0026] 1. Prepare millet materials: wolf tail, white quicksand, dead car 1, Baiqinzhou yellow, Laolaibian, red sticky valley, knife handle, Gougenhong, Qianchuanzi, big green seedlings, yellow sticky valley, red millet 15 kinds of millet materials, namely , yellow grain, duck beak, big grain yellow, and red seedling valley, were planted in the greenhouse; millet sprouts about 10d-15d after germination were taken, frozen in liquid nitrogen, and stored in a -80°C ultra-low temperature refrigerator for later use; 15 The information of the millet varieties is shown in Table 1.
[0027] Table 115 Millet Variety Information
[0028] serial number
[0029] 2. Millet DNA extraction: Extract the DNA of various millet materials with a plant genome kit, and use 1% agarose gel electrophoresis to detect the DNA purity; dilute the DNA concentration of each material to 50ng / μL, and place it at -20 ℃ in the refrigerator for later ...
Embodiment 2
[0051] The present embodiment provides a method for identifying millet varieties using SSR molecular marker technology, comprising the following steps:
[0052] 1. Prepare millet materials: plant 45 kinds of millet materials such as Gogenhong, Longclaw, Xiaobaigu, Qianchuan, Knife Handle, Dalihuang, Baishagu, etc. in the greenhouse; take millet sprouts about 10d-12d after germination, and liquid After freezing with nitrogen, put them in a -80°C ultra-low temperature freezer and store them for later use; the information of the 45 millet varieties is shown in Table 7.
[0053] Table 745 Millet Variety Information
[0054]
[0055] 2. Millet DNA extraction: Extract the DNA of various millet materials with a plant genome kit, and use 1% agarose gel electrophoresis to detect the DNA purity; dilute the DNA concentration of each material to 50ng / μL, and place it at -20 ℃ in the refrigerator for later use;
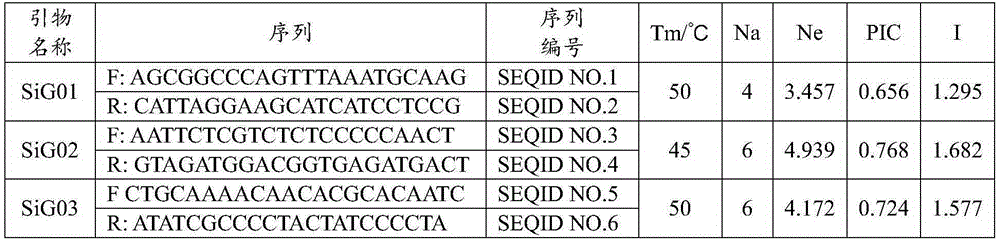
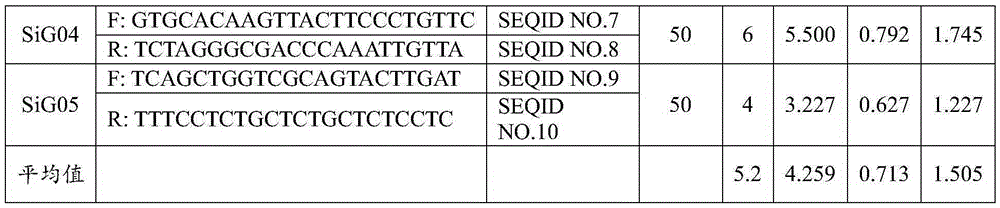
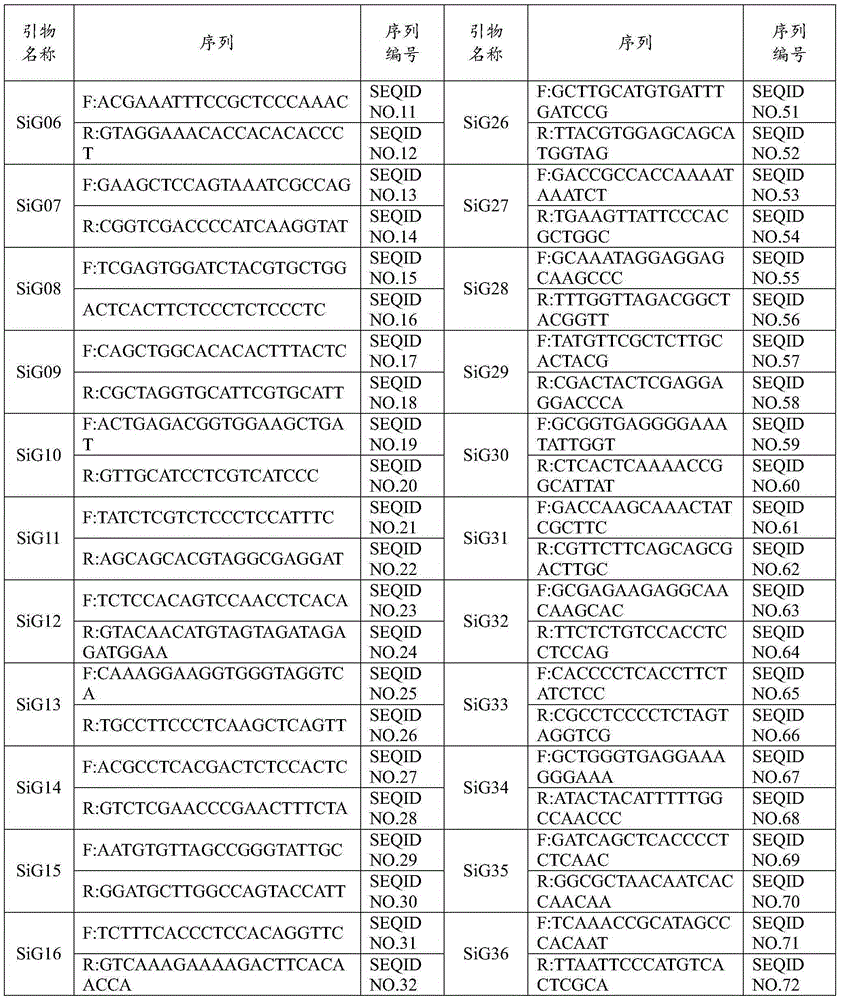
[0056] 3. SSR primers: 5 pairs of SSR core primers suitable for the iden...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com