Primer and probe for Zygosaccharomyces bailii detection and detection method thereof
A technology of Zygomyces beijerinckii and primer set, applied in the field of microbial detection, can solve the problems of undetectable damaged cells, low specificity, false positives, etc., to ensure consumer rights, high DNA purity, Guaranteed effect of specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1, the design of real-time fluorescent PCR detection method specific primer
[0041] Download the gene sequences of various microorganisms published in GenBank, and use DNAMAN software to compare and analyze, select the 26S ribosomal RNA gene sequence of Zygomyces beijerinferi to design specific primers and probes for fluorescent PCR (only Amplified DNA restriction fragments of Zygomyces baijerinferi, but not other microbial DNA restriction fragments), the primers and probes are as follows:
[0042] The upstream primer sequence is: 5'-TTTGATCAGACATGGTGTTTTGC-3';
[0043] The downstream primer sequence is: 5'-ATGCTGGCCCAGTGAACTG-3';
[0044] Probe sequence: 5'(FAM)-CCCCTCGCCTCTCGTGGGTG-3'(TAMRA).
Embodiment 2
[0045] Embodiment 2, the extraction of test sample DNA template
[0046] (2.1) Take 1mL sample and add it to a 1.5mL sterile centrifuge tube, centrifuge at 8000r / min for 5min, pour off the supernatant, add 1mL10mmol / LTris-HClpH8.0 to resuspend the precipitate, and spin at 8000r / min Centrifuge for 5 minutes, place it upside down on absorbent paper, and blot the liquid. Different samples should be blotted dry in different places on the absorbent paper;
[0047] (2.2) Yeast cell wall breaking: Add 600 μL of sorbitol buffer to the collected bacterial pellet, see Appendix A.2, add 50 U of Lyticase, shake well on a vortex shaker, and treat at 30°C for 30 minutes. Centrifuge at 4000r / min for 10min, discard the supernatant, and keep the precipitate;
[0048](2.3) Add 700 μL of 20g / LCTAB lysate to the cell pellet after breaking the wall, shake on a vortex shaker to mix well, and place in a 65°C water bath for 0.5-2 hours;
[0049] (2.4) Extraction: After cooling to room temperature, ...
Embodiment 3
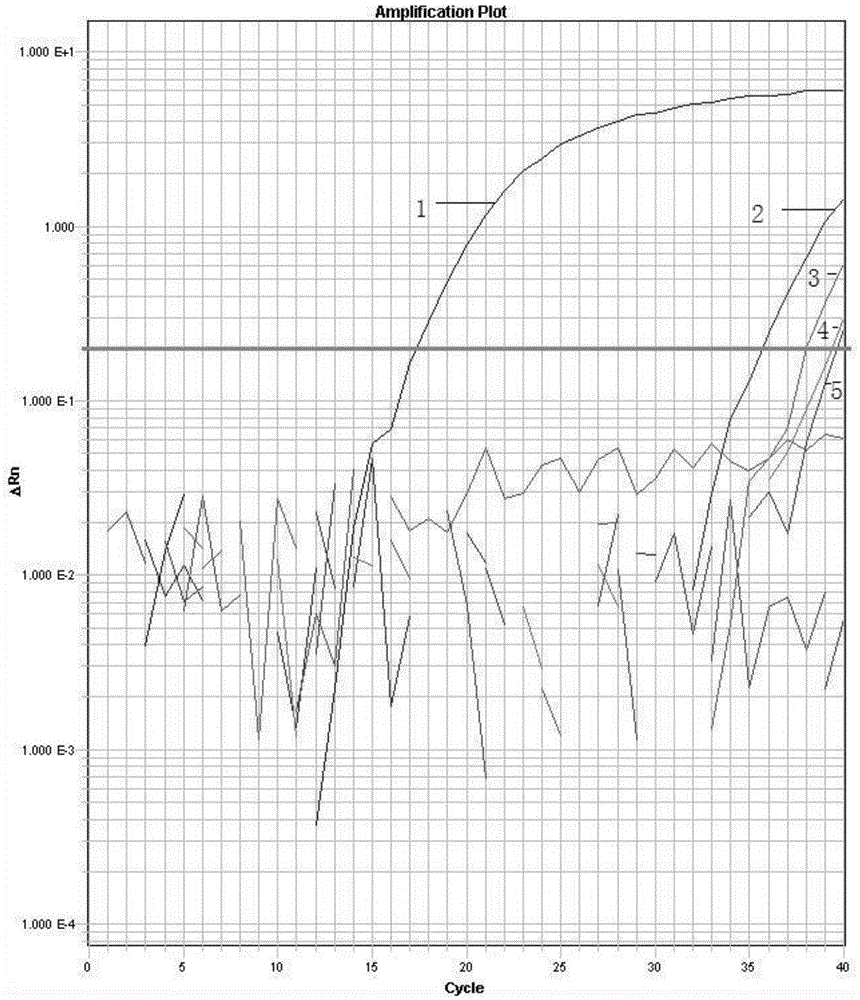
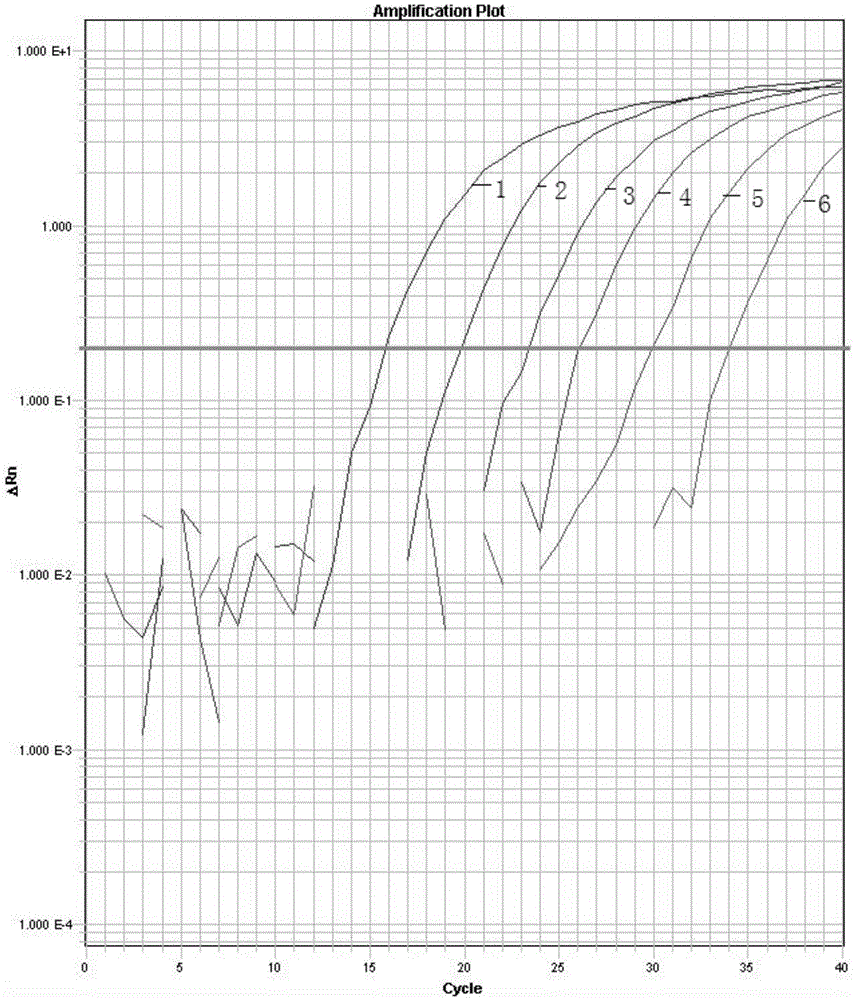
[0055] Embodiment 3, real-time fluorescent PCR detection of Zygomyces baijerin
[0056] Using the specific primers and probes designed in Example 1 above, PCR amplification was performed using the extracted DNA as a template.
[0057] (3.1) The real-time fluorescent PCR reaction system is 25 μL, including 12.5 μL 2×realtimePCRBuffer (containing dNTP, Mg 2+ and Taq enzyme), 1 μL of designed upstream primer and downstream primer (final concentration of 0.2 μM), 1 μL of probe (final concentration of 0.2 μM), 2 μL of extracted product DNA template, 7.5 μL of pure water;
[0058] (3.2) The PCR reaction program is: pre-denaturation: temperature 95°C, time 10 minutes, then PCR amplification: temperature 95°C, time 15 seconds, temperature 63°C, time 1 minute, cycle 40 times;
[0059] (3.3) Use the random software of the fluorescent quantitative PCR instrument to set the reaction parameters, save the file after the reaction, open the analysis software, and analyze the experimental res...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com