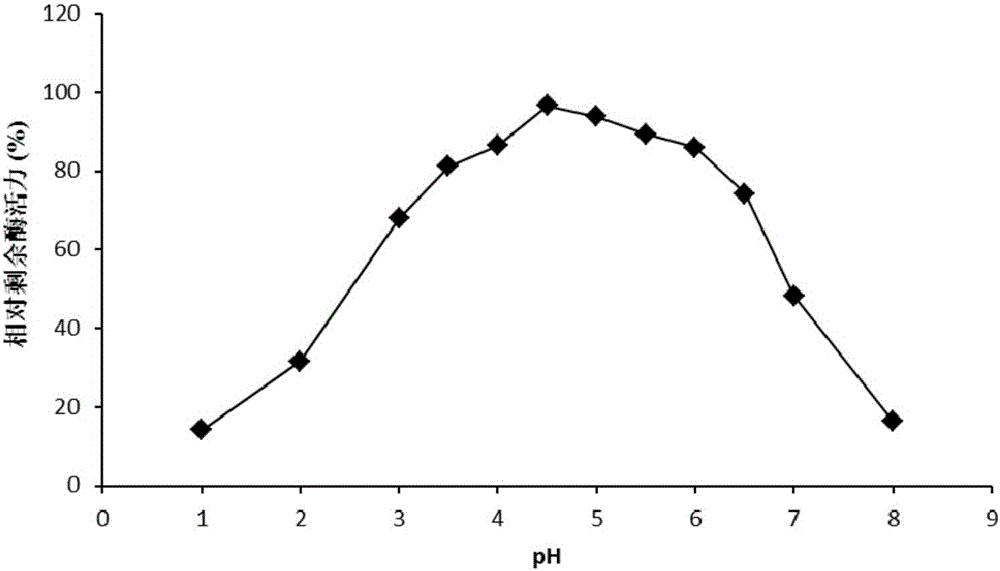
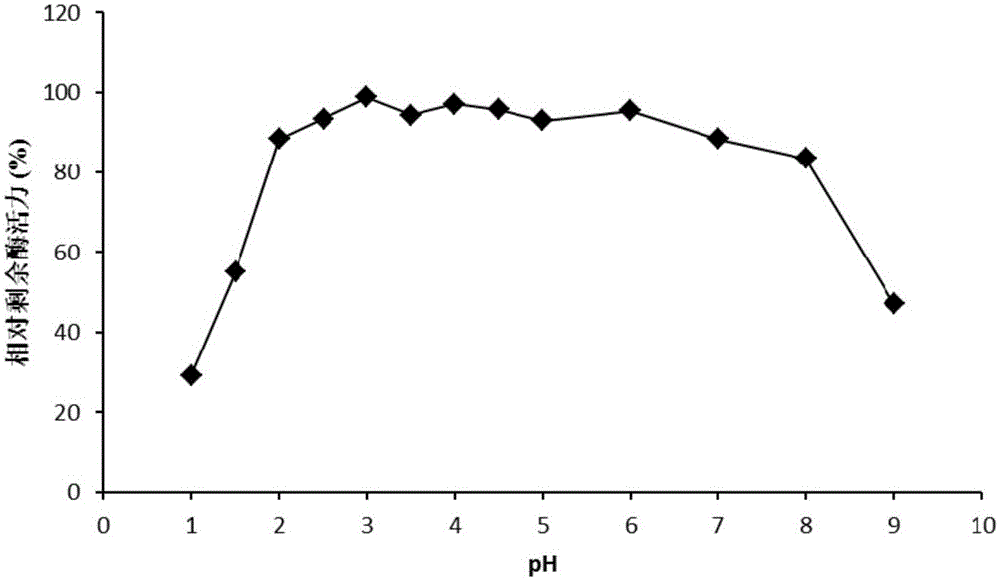
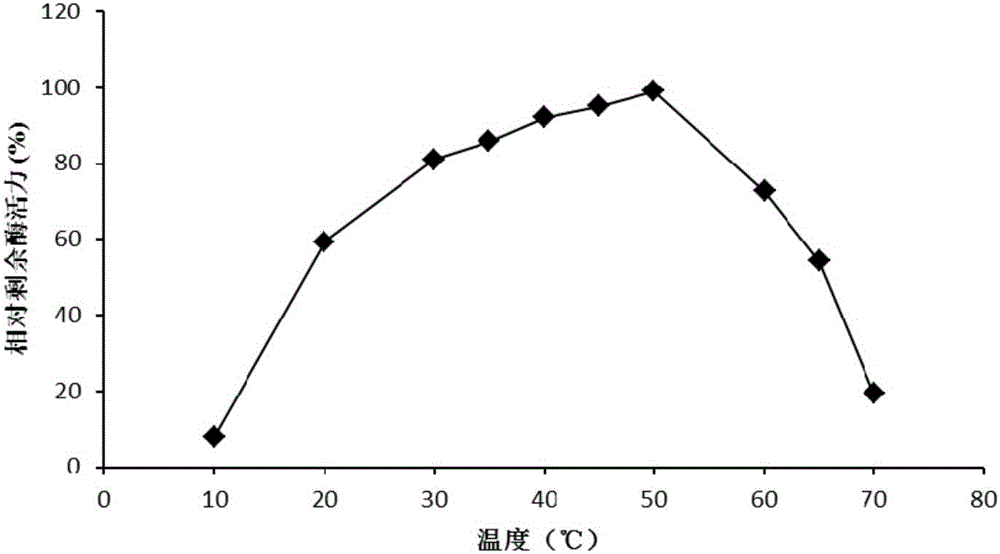
High temperature-resistant acid arabinfuranosidease AbfaHLB and gene and application thereof
A furanosidase and high temperature-resistant technology, applied in the field of genetic engineering, can solve the problems of insufficiency and poor thermal stability, and achieve the effects of good thermal stability and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Cloning of the Arabinofuranosidase Gene abfaHLB in Example 1 Bacillus badiusHLB
[0047] Extraction of Bacillusbadius HLB genomic DNA:
[0048] (1) Take 0.5-2mL culture liquid, centrifuge at 10000rpm for 30s, absorb the supernatant as much as possible, and collect the bacteria;
[0049] (2) Add 200 μL buffer RB to the EP tube to resuspend, centrifuge at 10,000 rpm for 30 s, and discard the supernatant;
[0050] (3) For Gram-positive bacteria: add 120 μL lysozyme, mix by inverting, and bathe in 37°C water bath for 30-60 minutes;
[0051] (4) Centrifuge at 12000rpm for 2min, discard the supernatant and resuspend the cells in 180μL buffer RB by shaking or pipetting;
[0052] (5) Add 20 μL of RNaseA (25 mg / mL) solution, vortex and mix, and place at room temperature for 5-10 min;
[0053] (6) Add 800 μL of binding solution CB, then add 100 μL of isopropanol, and immediately vortex to mix thoroughly, at this time flocculent precipitation may appear;
[0054] (7) Add the m...
Embodiment 2
[0065] The preparation of embodiment 2 arabinofuranosidase AbfaHLB
[0066] Design and synthesize expression primers according to the obtained gene sequence of arabinofuranosidase AbfaHLB:
[0067] P3: 5'-GATGAATTCATGTCTATGGATGTAGATCCACGTTTA-3';
[0068] P4: 5'-GCTGCGGCCGCTACATTTACACGTAAACGAATCCAAGA-3'.
[0069] Using the total DNA of Bacillus badius HLB as a template, PCR amplification was performed again to obtain the gene of arabinofuranosidase AbfaHLB with a recombination restriction site. The expression vector pPIC9 is subjected to double digestion (EcoRI+NotI), and the gene abfaHLB encoding arabinofuranosidase is double-digested (EcoRI+NotI) at the same time, and the gene fragment encoding mature arabinofuranosidase is digested and connected to the expression vector pPIC9 , obtain the recombinant plasmid pPIC-abfaHLB containing the Bacillusbadius HLB arabinofuranosidase gene abfaHLB and transform Pichia pastoris GS115 by electroporation to obtain the recombinant Pichia...
Embodiment 3
[0071] Example 3 Activity analysis of recombinant arabinofuranosidase AbfaHLB
[0072] Determination of the activity of arabinofuranosidase: the amount of the product p-nitrophenol generated by the enzymatic hydrolysis of the substrate pNPAf was measured at 405 nm. Reaction steps: Mix 250 μL 2mM pNPAf substrate with 150 μL buffer, add 100 μL appropriately diluted enzyme solution, react at 40°C for 10 min, add 1.5 mL 1M Na 2 CO 3 The reaction was terminated, and the OD value was measured at 405 nm.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com