Establishment method of gene library of fecal flora based on high-throughput gene sequencing
A fecal flora and gene sequencing technology, applied in the biological field, can solve the problems of low throughput, low efficiency, and cannot well meet the needs of scientific research and clinical practice, and achieve the effect of fast library construction and large throughput.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
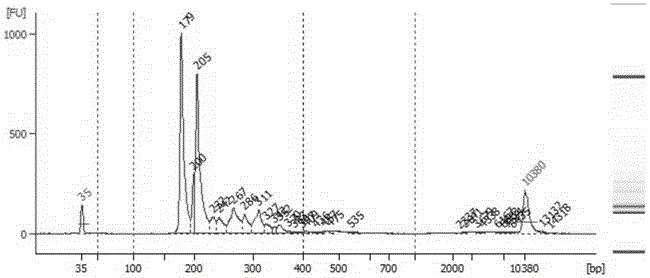
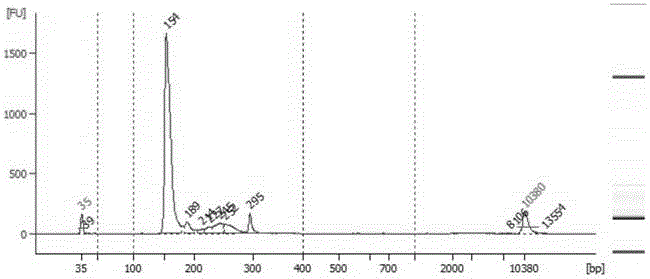
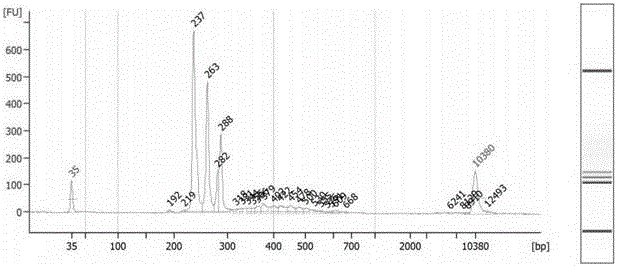
[0113] The technical solutions of the present invention will be described in further detail below in conjunction with the accompanying drawings and specific embodiments, but the present invention is not limited to the following embodiments. The materials used in the examples are the feces of a group of patients confirmed to be positive for Clostridium difficile AB toxin provided by the General Hospital of Guangzhou Military Region.
[0114] The specific steps of the experimental process for establishing the library of the present invention are as follows.
[0115] 1. Extract the total genomic DNA of fecal flora:
[0116]The total genomic DNA of the fecal flora was extracted strictly according to the operation manual of the TIANamp Stool DNA Kit (TIANGEN DP328), and the conditions were optimized for intestinal bacteria: the lysis temperature of the sample pretreatment was adjusted to 95°C and incubated for 10 minutes, so that the comparative Difficult-to-break Gram-positive ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com