A High-Throughput Screening Method for Prey Antagonists Based on Membrane-Bound Proteins and Fluorescence Complementation
A receptor antagonist and prey technology, applied in the field of genetic engineering, can solve the problems of insufficient use of compound libraries, poor reproducibility, and difficulty in ensuring activity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1. Vector construction and expression localization of CXCL12 ligand protein
[0073] Objective: To achieve the effect of localizing on the cell membrane by fused expression of CXCL12 gene with signal peptide gene and transmembrane peptide gene.
[0074] Plasmid construction: the mouse chemokine CXCL12 mature peptide coding sequence (shown in SEQ ID NO.5, specifically: KPVSLSYRC PCRFFESHIA RANVKHLKIL NTPNCALQIV ARLKNNNRQV CIDPKLKWIQ EYLEKALNK) was amplified and sequenced, and the secretory signal peptide sequence (from yeast Wbp1 signal peptide The sequence, shown in SEQ ID NO.1, is specifically:
[0075] MARVMRTDWNFFFCILLQAIFVVGTQTSRTLVLYSK) transmembrane peptide (from yeast Wbp1 transmembrane peptide sequence, shown in SEQ ID NO.2, specifically:
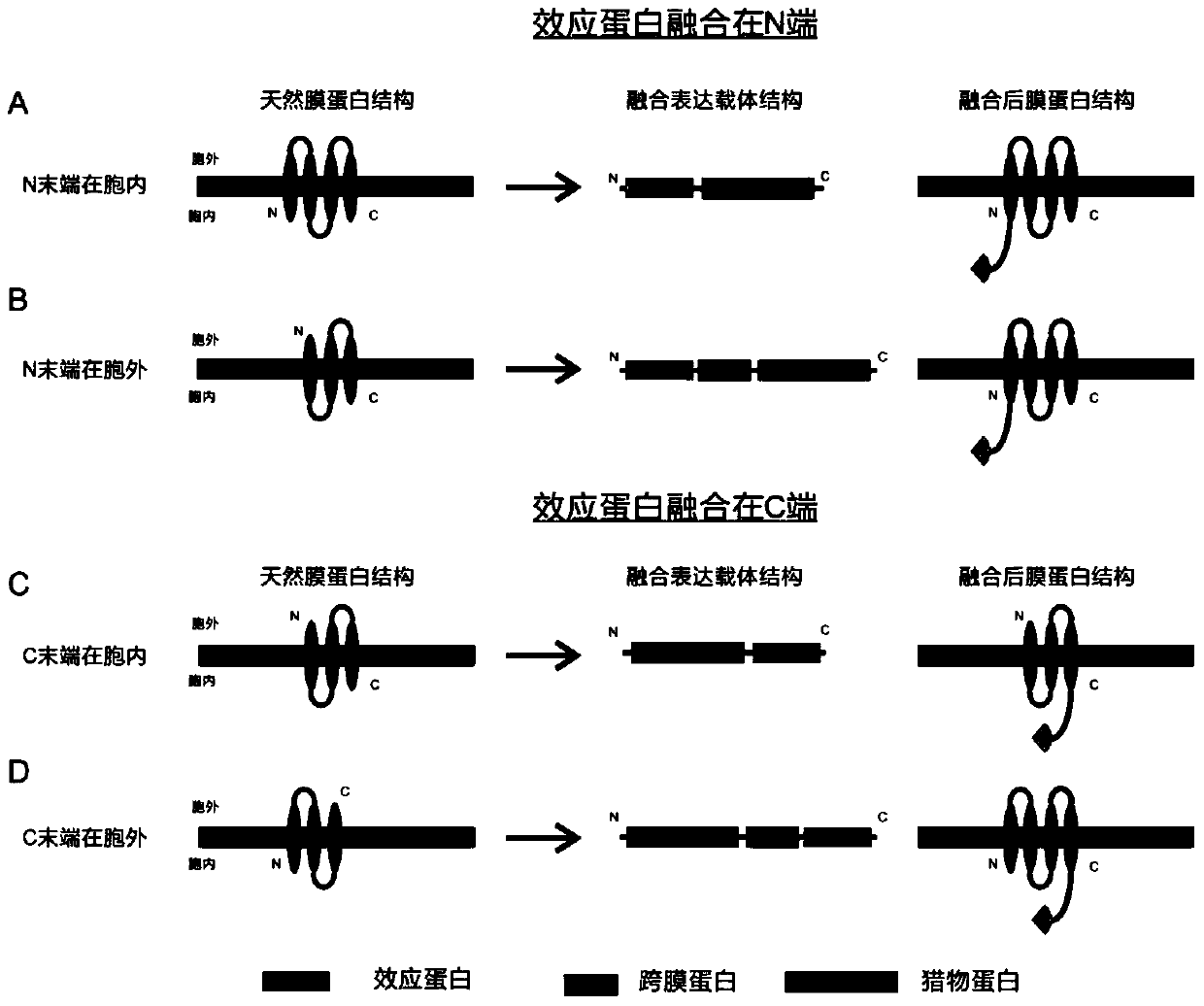
[0076] TGEFILPDRHGVFTFLTDYRKIGLSFTTDKDVKAIRHLANDEYPRSWEISNSWVYISAICGVIVAWIFFVVSFVTTSSVGKKLETFKKT) was fused, and then the green fluorescent protein (EGFP) reporter gene was connected, and the entire fusion protein wa...
Embodiment 2
[0080] Example 2. Construction and positioning of CXCR4 receptor protein vector
[0081] Objective: To clarify the expression localization of CXCR4 receptor in yeast cells.
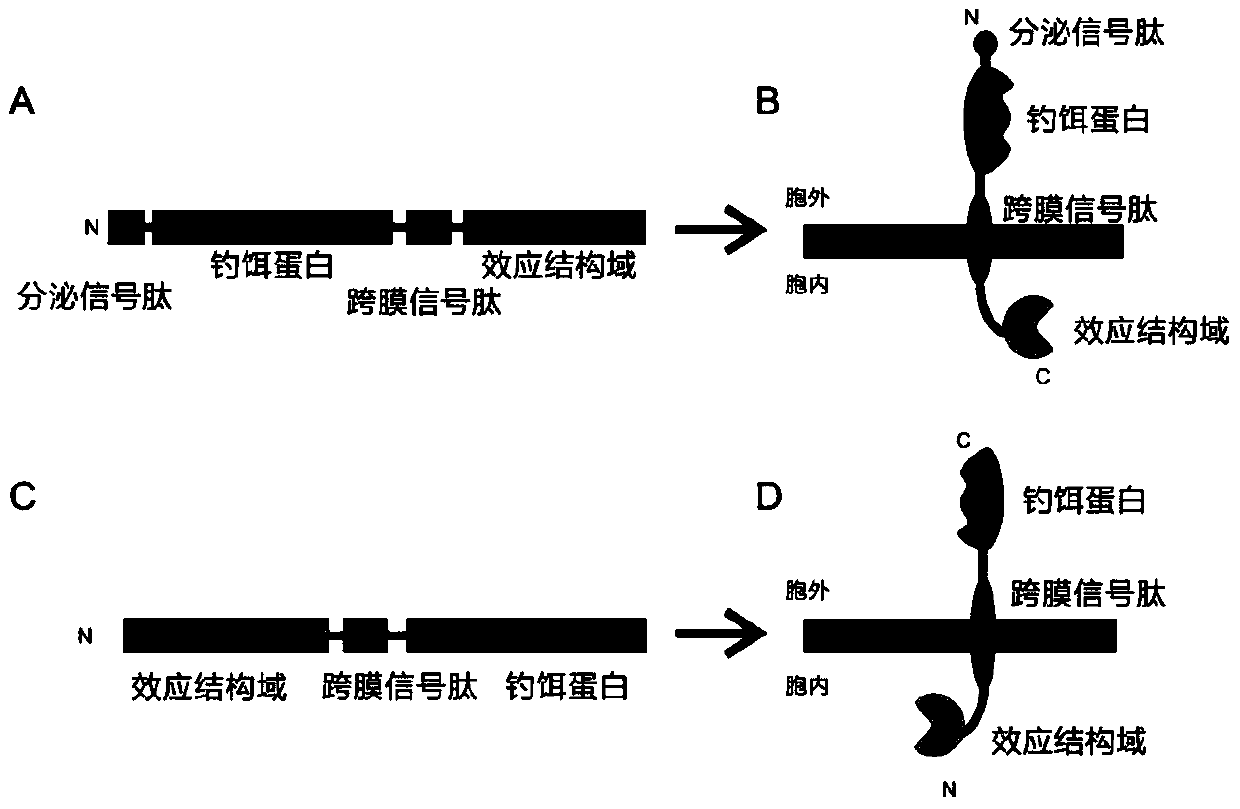
[0082] Vector construction: the CXCR4 receptor gene was cloned, fused with yellow fluorescent protein (YFP), and cloned into the yeast expression vector pGBK-T7 ( Image 6 A), denoted as: CXCR4-YFP plasmid. Among them, the relevant information of CXCR4 is detailed in:
[0083] CXCR4 Reference HTTP: / / WWW.UNIPROT.ORG / UNIPROT / P70658 ;(NP_034041.2)
[0084] Prediction of transmembrane structure of fusion protein: use TMHMM2.0 online analysis software to analyze the transmembrane topology of fusion protein. The software calculation results show that the fusion protein can correctly express the receptor protein to the cell membrane, and express the YFP protein fused to the C-terminus into the cell ( Image 6 B).
[0085] Plasmid transfection: transform the CXCR4-YFP plasmid into the Y187 cell line by LiA...
Embodiment 3
[0087] Example 3. Establishment of CXCL12-CXCR4 Interaction Detection Platform
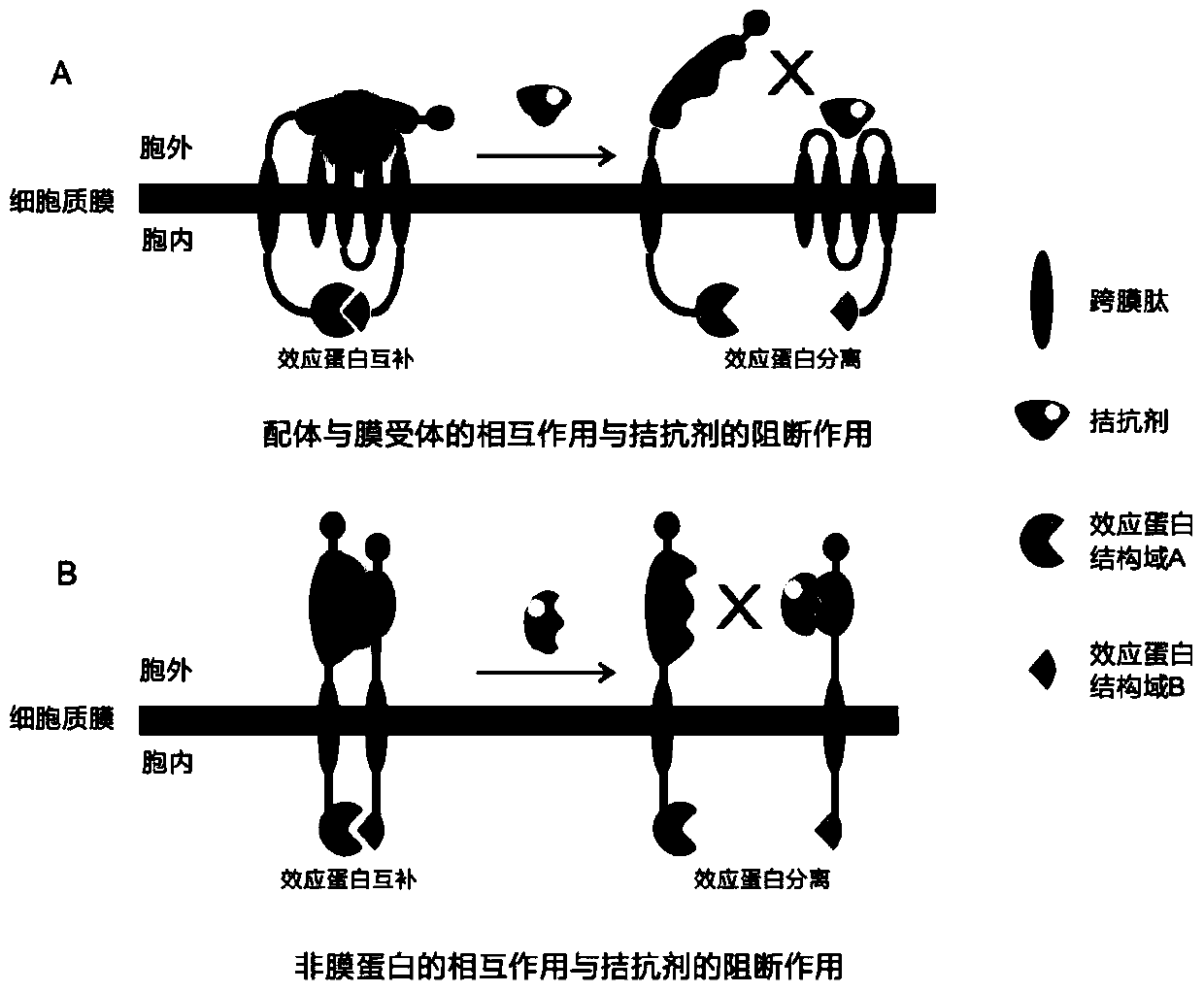
[0088] Example Purpose: To establish a detection platform for the interaction between CXCL12 and CXCR4 by bimolecular fluorescence complementation technology for subsequent screening of receptor antagonists.
[0089] Principle: The ligand gene CXCL12 (NP_038683.1) is fused with the C-terminal domain of yellow fluorescent protein (CYFP) through a transmembrane peptide, and the receptor CXCR4 (NP_034041.2) is fused with the N-terminal domain of yellow fluorescent protein (NYFP). Neither NYFP nor CYFP can fluoresce alone. Only when CXCL12 interacts with CXCR4, NYFP and CYFP undergo bimolecular fluorescence complementation (BiFC) and reconstitute into a complete active yellow fluorescent protein (YFP) with fluorescent function. The intensity of the yellow fluorescence (wavelength 529 nm) of the bacterial solution was detected by a fluorescent microplate reader.
[0090] Plasmid Construction: CXCL12-C...
PUM
| Property | Measurement | Unit |
|---|---|---|
| absorbance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com