Gene engineering arginine deiminase reformed through site directed mutagenesis
An arginine deiminase and site-directed mutagenesis technology, applied in the field of genetic engineering, can solve the problems of weak substrate affinity, low enzymatic activity, short in vivo half-life, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1: Construction of recombinant plasmid
[0029] Will Enterococcus faecalis SK32.001 was cultured to the mid-exponential growth stage, and 2 mL of the bacterial liquid was centrifuged at 10,000 r / min for 10 min to discard the supernatant. After lysozyme treatment for 30 min, genomic DNA was extracted according to the kit instructions.
[0030] The following primers were designed for the amplification of arcA:
[0031] FADI-2: 5'-CGCGGATCCA TGAGTCATCC AATTAATGT-3' (with Bam H I restriction site),
[0032] RADI-2: 5’-CCGCTCGAGT TAAAGATCTT CACGGT-3’ (with xho Ⅰ restriction site),
[0033] PCR amplification conditions: denaturation at 95°C for 3 minutes, 30 cycles (95°C for 30 s, 55°C for 30 s, 72°C for 210 s), and finally extension at 72°C for 2 min.
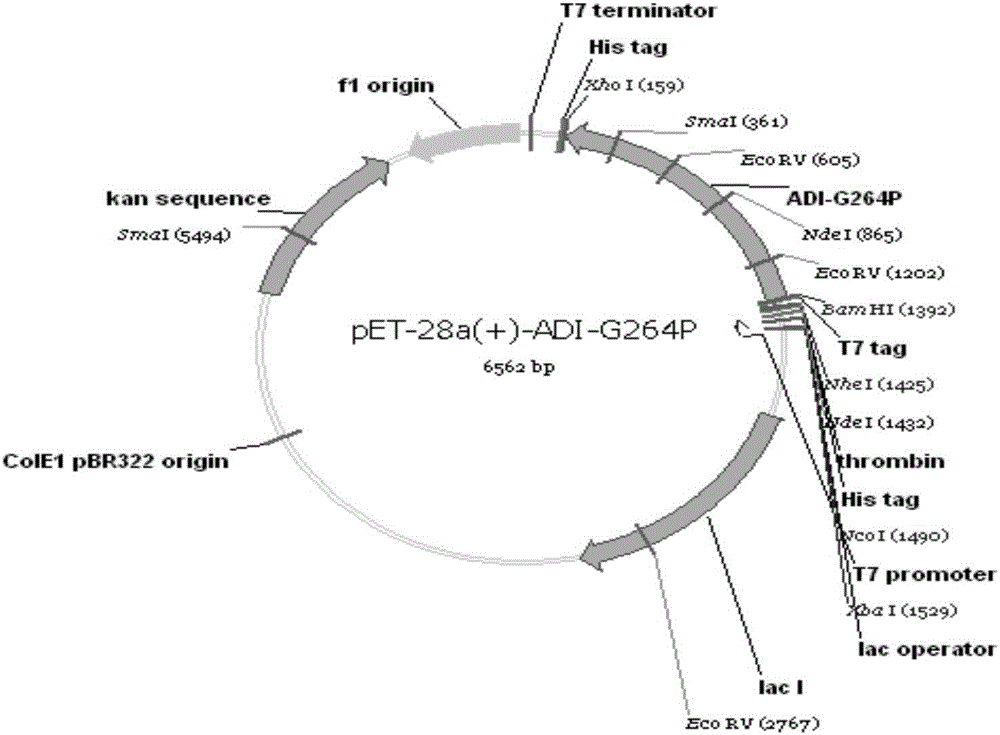
[0034] After purification of the amplified product, the Bam H I and xho Ⅰ Carry out double enzyme digestion on the PCR product and the vector pET-28a-c(+), recover the digested products separately, conne...
Embodiment 2
[0035] Example 2: Site-directed mutagenesis
[0036] according to Enterococcus faecalis Coded in SK23.001 arc The coding gene of A was designed for primers.
[0037] G264P-Forward primer: 5'-CTTGGCTTTT GATATC CCT G AACATCGTAA ATTC-3',
[0038] G264P-Reverse Primer: 5'-GATATCAAAA GCCAAGATAT TTTTGAATCC TA-3',
[0039] The underlined part represents the codon corresponding to the 264-position glycine encoded by the mutant gene. The PCR amplification system is:
[0040] 10×Reaction Buffer 5 μL dNTP mix 1 μL Forward primer (100 ng / μL) 1.25 μL Reverse primer (100 ng / μL) 1.25 μL Template pET-28a-ADI (10 ng) 2 μL pfu Turbo DNA polymerase (2.5U / μL)
[0041] After PCR amplification, add 1 μL to the reaction solution Dpn ⅠRestriction endonuclease (10 U / μL), incubate at 37°C for 1h to eliminate the template. Transform the PCR product into Escherichia coli DH5α cells, smear the plate, pick a single colony into LB medium, extract ...
Embodiment 3
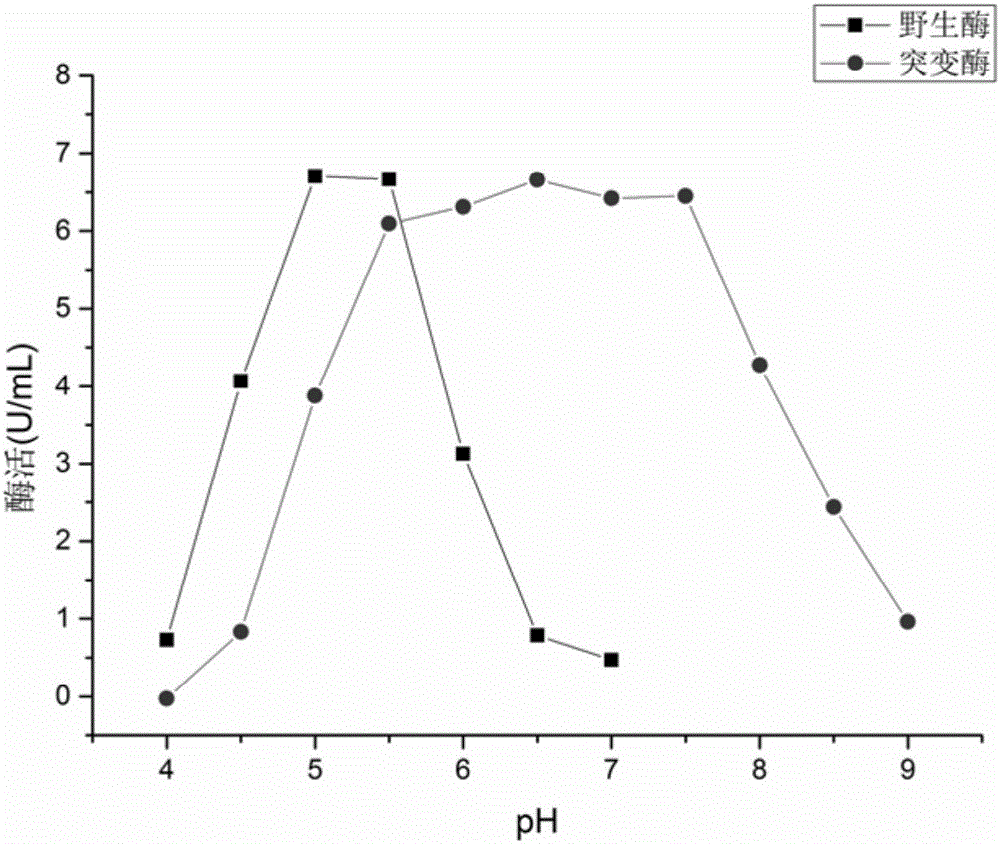
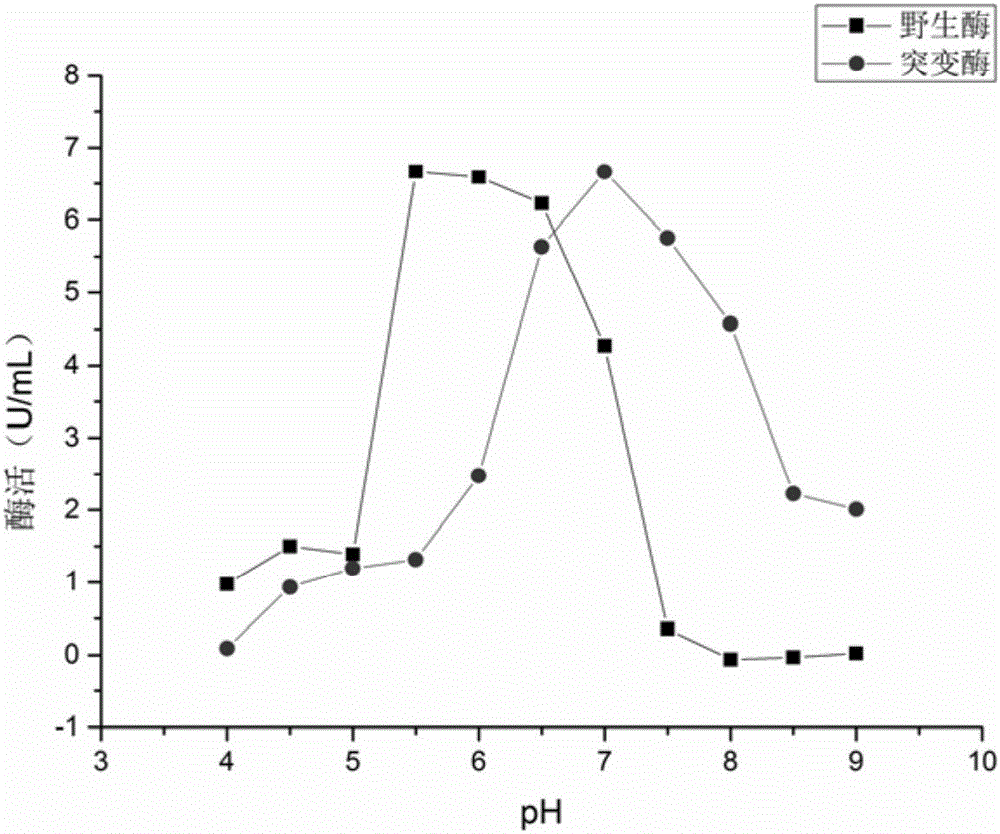
[0042] Example 3: Expression and purification of wild enzyme and mutant enzyme
[0043] Pick BL21(DE3) / pET-28a-ADI and pET-28a-ADI G264PSingle colonies were respectively cultured in LB medium containing 0.5mmol / L kanamycin at 37°C and 200r / min for 12h, then transferred to LB medium containing 0.5mmol / L kanamycin and incubated at 37°C , 200r / min culture to OD 600 In the range of 0.5-0.7, add 1 mmol / L IPTG to induce 9 h at 28°C and 200 r / min.
[0044] After the fermentation broth was centrifuged at 10000r / min and 4°C for 10min, discard the supernatant, wash twice with phosphate buffer, add 15-20mL of phosphate buffer to suspend the cells, and ultrasonically break for 15min (power 22W, break for 1s , intermittent 2s). Centrifuge at 4°C and 10,000 r / min for 10 minutes, collect the supernatant, which is the crude enzyme solution, and filter it with an aqueous membrane with a pore size of 0.22 μm.
[0045] Use Binding Buffer for Ni 2 + The chelating agarose resin column was pre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com