Carbonyl reductase, mutant and application thereof in preparation of antifungal drug intermediates
A technology of carbonyl reductase and amino acid, applied in the direction of oxidoreductase, application, enzyme, etc., can solve the problems of poor substrate tolerance, low production efficiency, low substrate tolerance concentration, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Example 1 Gene Cloning of Carbonyl Reductase SsCR
[0086] According to the open reading frame of carbonyl reductase SsCR, design upstream and downstream primers as follows:
[0087] Upstream primer SEQ ID No.3:
[0088] CCG GAATTC ATGACTACCTCAGTTTTCGT
[0089] Downstream primer SEQ ID No.4:
[0090] CCG CTCGAG TTAACCTTGTACCTTTCAAAA
[0091] Wherein, the underlined part of the upstream primer is the EcoR I restriction site, and the underlined part of the downstream primer is the Xho I restriction site.
[0092] The genomic DNA of xylose fermenting yeast CBS 6054 was used as a template for PCR amplification. PCR system: 2×TaqPCR MasterMix 25μl, upstream primer and downstream primer (10ng / μl) each 2.5μl, genomic DNA (100ng / μl) 1μl and ddH 2 O 19 μl. The PCR amplification program was: 95°C pre-denaturation for 5 minutes followed by 32 cycles of the following: denaturation at 94°C for 30 seconds, annealing at 50°C for 40 seconds, extension at 72°C for 1 minute, ...
Embodiment 2
[0093] Example 2 Preparation of Carbonyl Reductase Recombinant Expression Plasmid and Recombinant Expression Transformant
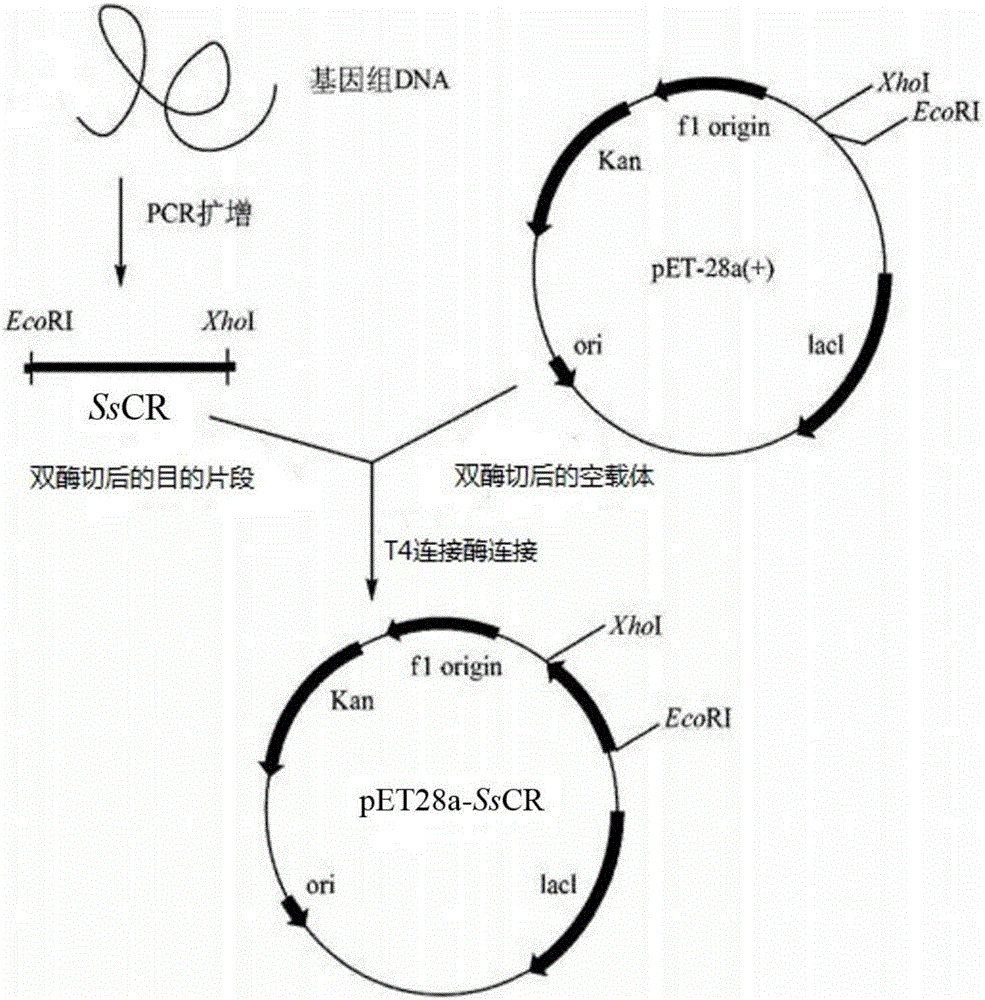
[0094] Such as figure 1 As shown, the carbonyl reductase target fragment obtained by PCR amplification in Example 1 and pET 28a empty plasmid were double-digested overnight with restriction endonucleases EcoR I and Xho I, then purified by agarose gel electrophoresis, and the DNA Kit recovery. The recovered target fragment and the empty plasmid vector were ligated under the action of T4 DNA ligase at 4°C for 12 hours to obtain the recombinant plasmid pET28a-SsCR.
[0095] Transform the resulting recombinant plasmid into E.coli DH 5α, spread it on an LB medium plate containing 50 μg / ml kanamycin, incubate at 37°C for 8 hours, perform colony PCR verification on the grown colonies, and pick colony PCR Positive clones with a target band of about 1000 bp in length were amplified. After verification by sequencing, the corresponding plasmids were extracted, fu...
Embodiment 3
[0096] Example 3 Carbonyl reductase SsCR mutant construction
[0097] The random mutation library of carbonyl reductase SsCR was constructed by error-prone PCR technique: pET28a_SsCR was used as template, For_EcoR I and Rev_Xho I were used as primers, and Taq DNA polymerase was used for error-prone PCR. To obtain a suitable mutation rate, a series of different MnCl 2 Concentration gradient (100μM~300μM MnCl 2 ) to build a mutation library. The PCR reaction conditions are as follows: in a PCR reaction system with a total volume of 50 μL, add 0.5-20 ng of template, 5 μL of 10×PCR buffer (Mg 2+ Plus), 5 μL dNTPs (2.0 mM each), 5 μL MnCl 2 (1mM), 2μL (10μM) of each pair of mutant primers, 0.5μL Taq DNApolymerase, add sterilized distilled water to 50μL. PCR reaction program: (1) denaturation at 95°C for 3 min; (2) denaturation at 94°C for 10 sec, (3) annealing at 60°C for 30 sec, (4) extension at 72°C for 90 sec, steps (2) to (4) were performed for 30 cycles in total, Finally,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com