Nanobody resisting to c-Myc labels
A nanobody and label technology, applied in the field of genetically engineered antibodies, single-domain heavy-chain antibodies or peptides, can solve the problems of limited sources of polyclonal antibodies, cumbersome and complicated process of R&D and production of monoclonal antibodies, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Construction of an immune library of anti-c-Myc tag single domain heavy chain antibody (ie single domain heavy chain antibody against c-Myc tag)
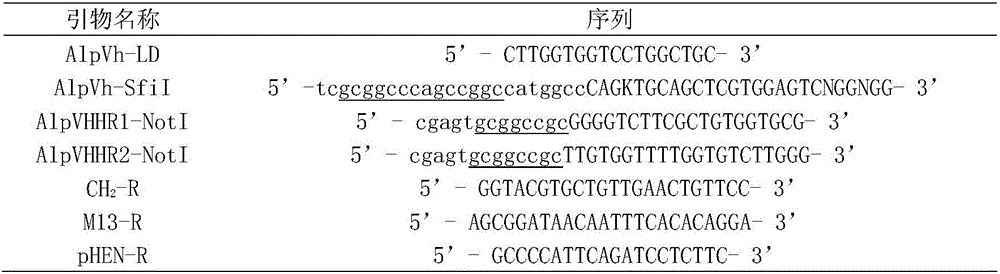
[0024] The c-Myc tag was covalently coupled to bovine serum albumin (BSA) to obtain the c-Myc artificial antigen c-Myc-BSA. After emulsifying 300 μg of c-Myc-BSA with Freund's complete adjuvant, Alpacas (Lama pacos) were immunized by subcutaneous multipoint injection. For booster immunization, 150 μg c-Myc-BSA was emulsified with Freund's incomplete adjuvant at intervals of 2 weeks. Blood was collected from the vein 7 days after each immunization, and the serum titer was determined by indirect ELISA method. The sample with the highest serum titer was selected to separate lymphocytes. cells, RNA was extracted.
[0025] The extraction of RNA was carried out according to the instruction manual of RNAiso reagent from TAKARA company. Using RNA as a template and oligo dT as a primer, the first strand of cDNA was synthesized accor...
Embodiment 2
[0034] Panning and Identification of Anti-c-Myc Tag Single Domain Heavy Chain Antibody
[0035]The single domain heavy chain antibody against c-Myc tag was panned from the anti-c-Myc tag single domain heavy chain antibody immune library obtained in Example 1 by solid phase affinity panning. Add 120 μL of Myc-GST fusion protein (a protein fused with Myc tag and glutathione) diluted with PBS to each enzyme-labeled well, and coat overnight at 4°C. The coating concentration for each round of panning is 100, 75, 50 μg / mL; aspirate the coating solution, wash the plate 5 times with PBS, add 300 μL 3% BSA-PBS to each well, block for 2 hours at 37°C; wash the plate 5 times with PBS, add 100 μL phage antibody library (about 1×10 11 CFU), 37°C, incubate for 2.0 h; aspirate unbound phage, wash the plate with PBST (containing 0.5% Tween-20) for 3-5 times (increase 5 times for each round), and then wash the plate with PBS for 15-25 times; Use 100 μL eluent (glycine-hydrochloric acid, pH 2....
Embodiment 3
[0045] Scale Production of Anti-c-Myc Tag Single Domain Heavy Chain Antibody
[0046] Obtaining the DNA fragment encoding the anti-c-Myc tag single-domain heavy-chain antibody: 1. Using restriction endonuclease SfiI / NotI, double-digest the phagemid pHEN-anti-c-Myc single-domain heavy-chain antibody gene, and agar Glycogel electrophoresis to recover the anti-c-Myc tag single domain heavy chain antibody gene; 2. Directly send the anti-c-Myc tag single domain heavy chain antibody coding sequence to a biotechnology service company for chemical synthesis; 3. Design specific primers, through PCR technology amplifies from a cDNA library derived from alpaca (Lama pacos).
[0047] The obtained anti-c-Myc tag single domain heavy chain antibody gene fragment was cloned into the expression vector pET25-flag (the c-Myc tag carried by the vector itself was replaced with the Flag tag: DYKDDDDK), identified by PCR and enzyme digestion, and constructed Complete the E. coli expression plasmid ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com