Plasmid and cell for promoting biotin synthesis and method for promoting synthesis of biotin
A biotin and enzyme synthesis technology, applied in the field of metabolic engineering, can solve the problem of low yield and achieve the effect of improving biotin synthesis efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Related primer design
[0035] According to the published biotin operon gene sequence of Bacillus subtilis in Genebank gi:2627063, primers Bio-EcoRI-up, Bio-XhoI-down and pET-EcoRI-up, pET-NotI-down were designed respectively. According to the Saccharomyces cerevisiae sam2 gene sequence published in Genebank gi:852113, design primers
[0036] pET-EcoRV-up, pET-XhoI-down. According to the Pseudomonas gene sequence published in Genebank gi:1002825811, two pairs of primers pACYC-EcoRV-up, PACYC-XhoI-down and pACYC-EcoRI-up, PACYC-NotI-down were designed respectively. The primer design results are shown in the table 1.
[0037] Table 1 The primers involved in the present invention
[0038] Primer
Embodiment 2
[0039] Example 2: Construction of recombinant plasmid pACYCDuet-bioBFHCD-bioA
[0040] The bioA (1407bp) and bioBFHCD (4489bp) genes were amplified by pACYC-EcoRV up, PACYC-XhoI-down and pACYC-EcoRI-up, PACYC-NotI-down using the P.putida KT2440 genome as a template. The pACYCDuet-1 plasmid and the bioBFHCD fragment were digested with EcoRI and NotI, and then ligated to the multiple cloning site (mcsl) behind T7promoler-1 to obtain the recombinant plasmid pACYCDuet-bioBFHCD.
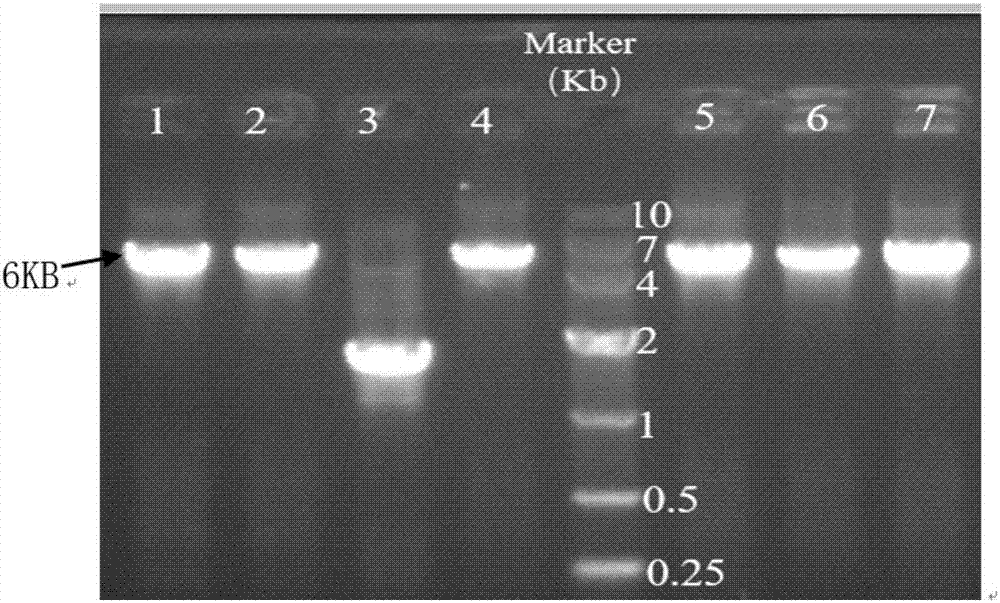
[0041] Transform the recombinant plasmid pACYCDuet-bioBFHCD into the cloning host DH5a, culture overnight, extract the recombinant plasmid, digest the pACYCDuet-bioBFHCD recombinant plasmid and bioA fragment with EcoRV and XhoI, and enzyme-link to the multi-cloning site behind T7promoler-2 Click on mcs2 to obtain the recombinant plasmid pACYCDuet-bioBFHCD-bioA, the electrophoresis of the recombinant plasmid is as follows figure 1 .
Embodiment 3
[0042] Embodiment 3: Construction of recombinant plasmid pETDuet-bioW-sam2
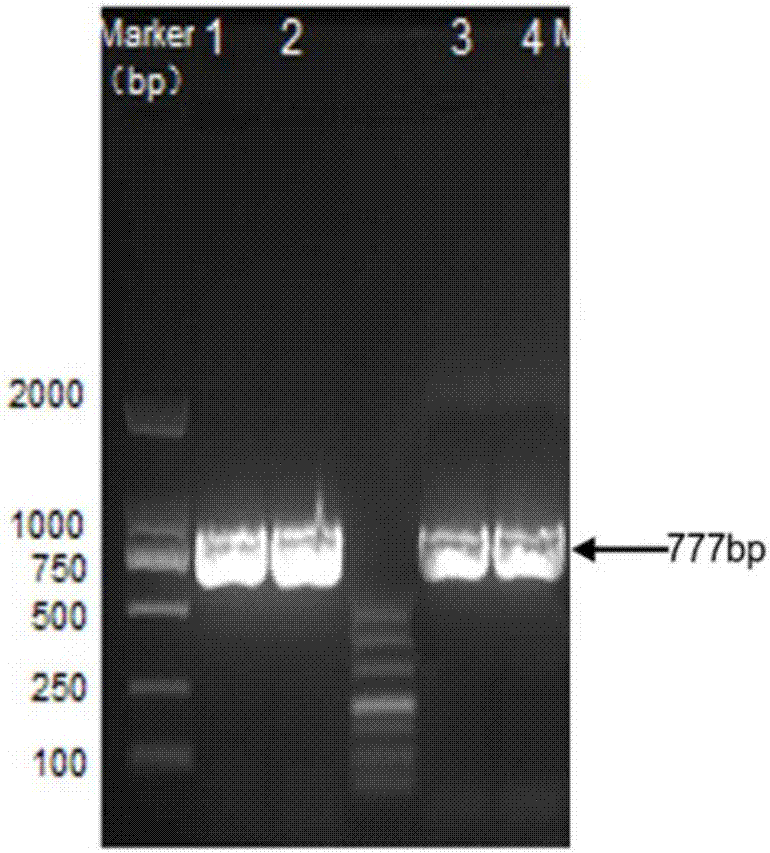
[0043] Through primers pET-EcoRI-up and pET-NotI-down, using B. subtilis168 genomic DNA as template, carry out bioW (777bp) gene amplification, the amplification result is as follows figure 2 , Purified by PCR, pETDuet-1 plasmid and bioW fragments were digested with EcoRI and NotI, and enzyme-linked to the multiple cloning site mcs1 behind T7promoler-1 to obtain the recombinant plasmid pETDuet-bioW.
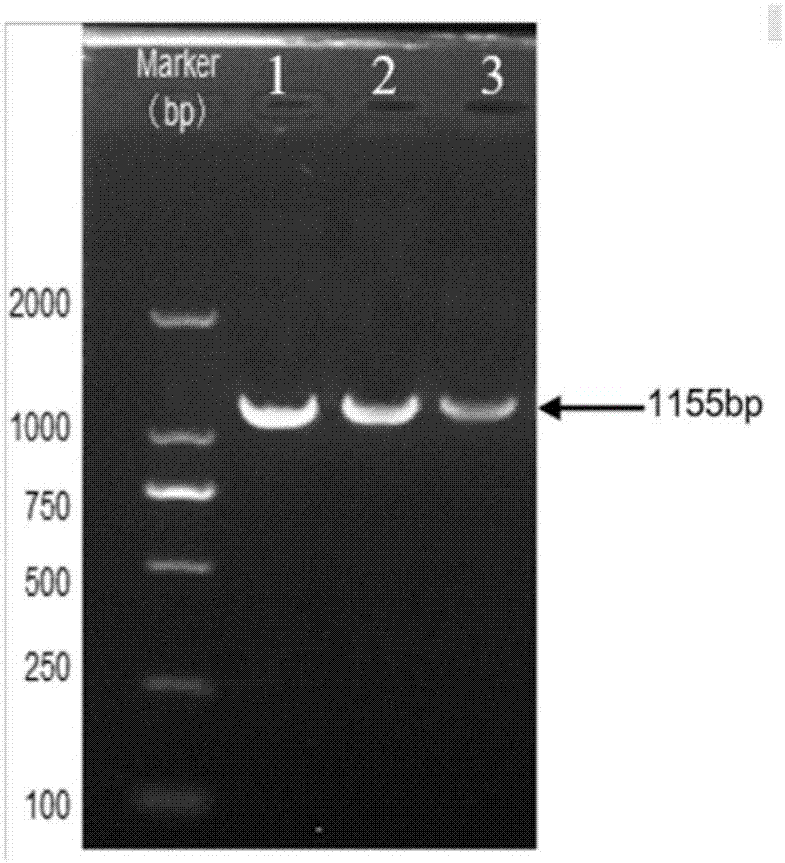
[0044] The recombinant plasmid pETDuet-bioW was transformed into the cloning host DH5a, cultivated overnight, and the recombinant plasmid was extracted. Using S.cerevisiae ZJU001 genomic DNA as a template, using pET-EcoRV-up and pET-XhoI-down primers to amplify the sam2 gene (1155bp), the electrophoresis of the amplified product is as follows image 3 . After double-digesting the pETDuet-bioW recombinant plasmid and sam2 gene fragment with restriction endonucleases EcoRV and XhoI, use T 4 The DNA lig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com