Specific gene segment suitable for real-time fluorescent quantitative PCR detection of pseudomonas PPZ-1 as well as primers thereof and applications
A real-time fluorescence quantification, Pseudomonas technology for DNA/RNA fragments, microbe-based methods, microbial assay/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Screening and primer design of embodiment 1 specific gene sequence
[0034] According to the genome sequencing results of the strain PPZ-1, the coding genes were compared in major general functional databases, and the unannotated gene sequences were screened. These sequences are the possible specific gene sequences of the strain; the sequences are respectively shown as SEQ ID Shown in NO.4, SEQ ID NO.1, SEQ ID NO.5 and SEQ ID NO.6.
[0035] Primers were designed respectively for the above gene sequences, and the primer of SEQ ID NO.4 was recorded as primer set 1, and the nucleotide sequence was as follows:
[0036] F: TCTCCGCCGCCCAGATAA; R: TGGATGTCCCGTTGAAGC
[0037] The primer of SEQ ID NO.1 is recorded as primer set 2, and the nucleotide sequence is as follows:
[0038] F: GCGGGAGGTGTTGCTGTT; R: CCTATTGCCCTTGCGTTC
[0039] The primer of SEQ ID NO.5 is recorded as primer set 3, and the nucleotide sequence is as follows:
[0040] F: CGGGTTATGGGCAGCAGA; R: GGTTGGCAA...
Embodiment 2
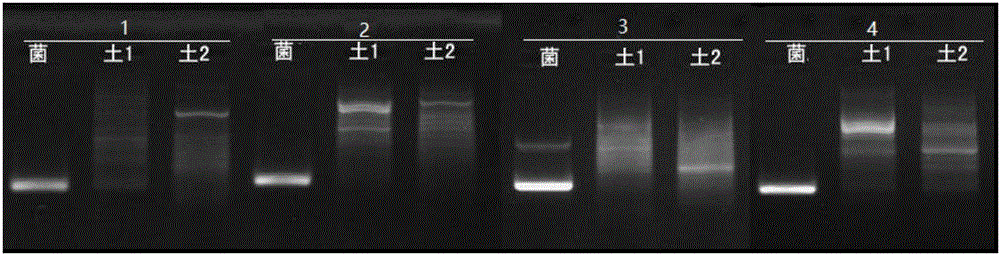
[0044] The above primers were used to amplify Pseudomonas (Psedomonassp.) PPZ-1 and two kinds of polycyclic aromatic hydrocarbon contaminated soil sample DNA by PCR, and the amplified products were observed by agarose gel electrophoresis.
[0045] Genomic DNA was extracted using a soil genomic DNA extraction kit to prepare template DNA.
[0046] The PCR amplification system is as follows, the total system is 25 μl:
[0047] Template DNA 0.5μl; Primer F (10μM) 0.5μl; Primer R (10μM) 0.5μl; dNTP (10mM) 0.5μl; TaqBuffer (10×) 2.5μl; MgCl 2 (25mM) 2μl; Taq enzyme (5U / μl) 0.2μl; H 2 O 18.3 μl;
[0048] The PCR reaction conditions were: pre-denaturation at 95°C for 3 min; denaturation at 94°C for 30 s, annealing at 57°C for 30 s, extension at 72°C for 30 s, 35 cycles; repair extension at 72°C for 8 min.
[0049] Electrophoresis conditions are: 1.5wt% agarose gel, 1×TAE, 150V, 100mA, 20min.
[0050] The result is as figure 1 As shown, 1, 2, 3, and 4 correspond to the above prime...
Embodiment 3
[0052] Example 3 Cloning and sequencing
[0053] Cut off the target band with a scalpel, recover it with the kit, connect it with the T carrier, transform it into Escherichia coli competent cells, spread it on an ampicillin-resistant plate, and culture it at 37°C for 24 hours. The clone was cultured in liquid LB medium for 24 hours, and the plasmid was extracted; PCR amplification was performed using the plasmid as a template to obtain a positive target plasmid. After the plasmid was sequenced and identified correctly, the OD of the plasmid was measured with an ultraviolet spectrophotometer 260 The value of is converted into copy number by formula.
[0054] The specific calculation is as follows:
[0055] (1): Plasmid concentration conversion formula (copies / μl)=(mol number / μl)×6.02×10 23 =[mass (g) / molecular weight] / μl×6.02×10 23 = [mass (ng) x 10 -9 / molecular weight] / μl×6.02×10 23 =Concentration (ng / μl)×6.02×10 14 / molecular weight
[0056] (2): Molecular weight = (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com