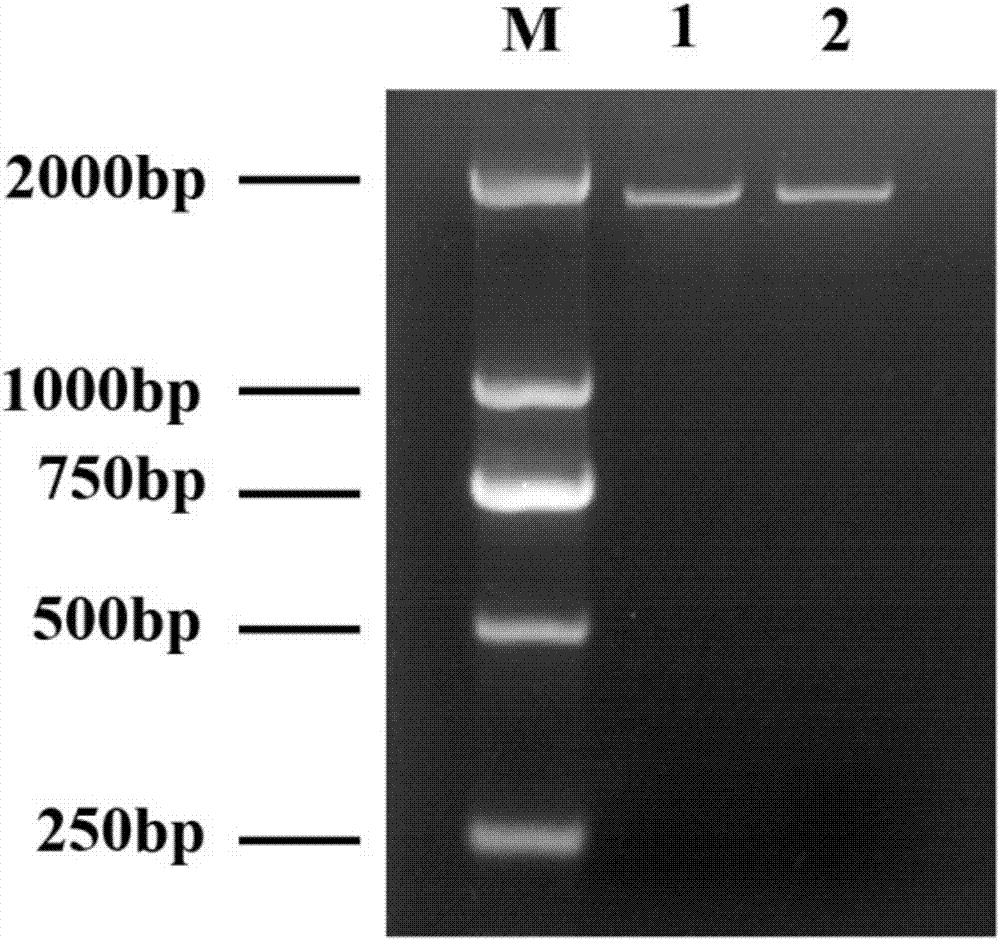
Molecular identification method of HMGR gene mRNA in pear stock-scion transfer
A technology for molecular identification and gene transfer, applied in DNA/RNA fragments, genetic engineering, plant genetic improvement, etc., can solve problems such as high cost, time-consuming, and differences, and achieve high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1 test tube micro-grafting
[0040] Select rootstock Pyrus betulaefolia Bunge and scion pear (Pyrusbretschneideri) tissue culture seedling young leaves, constant light source, light intensity 1500lx, light cycle 14h / 10h day and night, room temperature 23~25℃, relative humidity 85%, 30~40d Succession once.
[0041] Table 1 Ya pear and Du pear tissue culture medium
[0042]
[0043] After liquid nitrogen treatment, they were stored in a -80°C refrigerator for gene cloning. Under aseptic conditions, remove the head of the pear tissue-cultured seedlings after 30 days of propagation, leave about 1.5cm long stem section as a rootstock, and cut about 0.5cm hole longitudinally along the top; take the Yali tissue-cultured seedlings with a seedling height of 1cm, Leave 2-3 leaves at the top, cut the base into a "V" shape, insert it into the cut of the rootstock, wrap it with tin foil and fix it, and put it into the medium MS+0.5mg·L –1 6-BA+0.1mg·L –1 Grown in ...
Embodiment 2
[0044] Example 2 Gene RNA Extraction
[0045] Total RNA was extracted from plant materials using the CTAB method (Zhang Yugang et al., 2005). Total RNA was extracted from the leaves and stems of Du pears, Ya pears, rootstocks and scions after grafting, dissolved in 30 μL DEPC water, and placed at -80 for electrophoresis detection. ℃ refrigerator for future use.
[0046] (1) Remove DNA from RNA:
[0047] Add ingredients and reaction system as follows:
[0048]
[0049] (2) Treat at 37°C for 30 minutes; add 550 μL RNase-free water (treated with 0.1% DEPC), then add an equal volume of 600 μL CI and mix well;
[0050] (3) Centrifuge at 10000rpm for 10min at 4°C, absorb the supernatant, then add an equal volume of CI and mix gently; centrifuge at 10000rpm for 10min at 4°C;
[0051] (4) Aspirate the supernatant, add 2 times the volume of absolute ethanol to the tube, and precipitate at -20°C for 1 hour;
[0052] (5) Centrifuge at 12000rpm for 20min at 4°C; discard the superna...
Embodiment 3
[0055] Example 3 reverse transcription of RNA into cDNA
[0056] The reverse transcription system and procedures are as follows:
[0057] (1) Add the following components to a 0.2mL RNase-free PCR tube on ice:
[0058]
[0059] (2) Centrifuge briefly after mixing, put in a PCR instrument at 70°C for 10 minutes, and then immediately put it on ice for 5 minutes;
[0060] (3) Continue to add the following components to the PCR tube in the previous step on ice:
[0061]
[0062] (4) Instantaneous centrifugation after mixing, put into a PCR machine at 42°C for 60 minutes, and 70°C for 10 minutes. After the reaction is completed, take out the product and store it at -20°C for later use. The obtained cDNA was used as a template for PCR amplification described below.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com