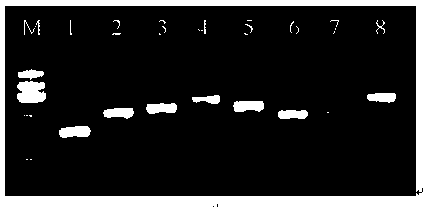
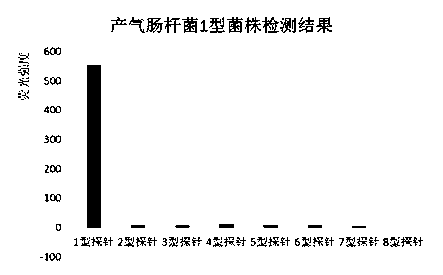
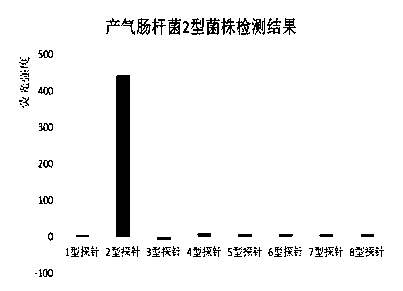
Suspension array for detecting serotype enterobacter aerogenes typing and application
A technology of Enterobacter aerogenes and serotyping, applied in the determination/inspection of microorganisms, resistance to vector-borne diseases, DNA/RNA fragments, etc., can solve the problem of incomplete antiserum, long cycle, difficulties in antiserum production and quality control, etc. problem, to achieve the effect of high detection throughput and short detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Design and Preparation of Specific Primers for Enterobacter aerogenes Serotype 1-8
[0069] (1) Both Enterobacter aerogenes serotypes 1-3 and 5-8 were selected wzy The gene is the target gene sequence, and the glycosyltransferase gene is selected for serotype 4 orf8 is the target gene sequence.
[0070] (2) Import the selected target gene sequences for Enterobacter aerogenes serotypes 1-8 into the primer design software Primer Primer 5.0, and set parameters. Among them, select the sense strand and complementary strand output mode; the sequence amplification length is 150-350bp; Haripin: none; Dimer: none: False Priming: none; Cross Dimer: none. Run the program to obtain a pair of specific primer sequences for the sense strand and the antisense strand of each serotype.
[0071] (3) Send the designed primer sequences to Thermo Fisher Scientific (China) Co., Ltd. for DNA synthesis and PAGE purification for future use. Among them, the synthesis of the antisense strand...
Embodiment 2
[0073] Design and Preparation of Specific Probes for Enterobacter aerogenes Serotype 1-8
[0074] (1) Both Enterobacter aerogenes serotypes 1-3 and 5-8 were selected wzy The gene is the target gene sequence, and the glycosyltransferase gene is selected for serotype 4 orf8 is the target gene sequence.
[0075] (2) Import the selected target gene sequences for Enterobacter aerogenes serotypes 1-8 into the primer design software Primer Primer 5.0, and set parameters. Wherein, only the sense strand output mode is selected; Haripin: None; Dimer: None: False Priming: None; Cross Dimer: None, and the position of the sequence is within the position of the sense strand and antisense strand primers in Example 1. Run the program to get 1 specific probe for each serotype.
[0076] (3) Send the designed probe sequence to Thermo Fisher Scientific (China) Co., Ltd. for DNA synthesis. At the same time, 12 carbon atoms are connected to the 5' end of the sequence as a connecting arm, and t...
Embodiment 3
[0078] Coupling of specific probes to microspheres (need to be protected from light)
[0079] (1) Suspend the microspheres on the vortex at the highest speed for 30 s, check the numbers of the microspheres and probes, and mark them.
[0080] (2) Take 80 microliters of microspheres in a 1.5mL centrifuge tube, centrifuge at 12000 rpm for 2 minutes.
[0081] (3) Discard the supernatant, resuspend with 10 microliters of 0.1 mol / L 2-(N-morpholine)ethanesulfonic acid solution (MES) (pH4.5), and vortex thoroughly to disperse the microspheres;
[0082] (4) Add 2 μl of probe (placed at room temperature in advance) and 6 μl of freshly prepared 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide salt at a concentration of 10 mg / mL salt solution (EDC), mix well, and incubate at room temperature in the dark for 30 minutes (shake and mix every 15 minutes).
[0083] (5) Add 6 microliters of freshly prepared 10 mg / mL EDC solution, mix well, and incubate at room temperature in the dark for 30 minu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com