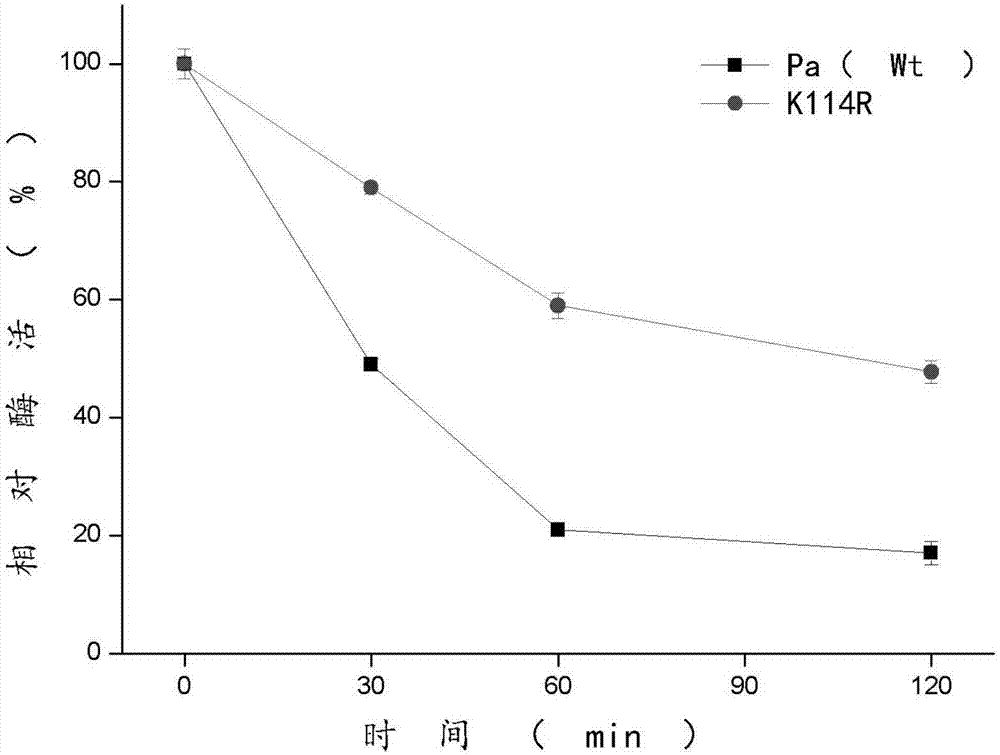
Phenylalanine amino mutase mutant derived from pantoea agglomerans
A technology of phenylalanine amino and mutase, which is applied in the field of enzyme engineering, can solve problems such as unsatisfactory expression, and achieve the effects of improved thermal stability, good pH stability, and good enzymatic properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] (1) Construction of mutant K114R:
[0035] The Pa-PAM gene (as shown in SEQ ID NO.1) was synthesized by chemical synthesis, and the gene was cloned at the NdeI and Hind III restriction sites of the pET28a plasmid, which was completed by Tianlin Biotechnology Company to obtain pET28a-PAM recombinant plasmid. Using pET28a-PAM as a template, use the primers shown in Table 1 to perform PCR under the conditions shown in Table 2, and transform the PCR product into E. coli JM109 to obtain the recombinant plasmid pET28a-PAM-K114R carrying the gene encoding the mutant. The recombinant plasmid pET28a-PAM-K114R was transformed into E. coli BL21 strain to obtain the recombinant strain BL21 / pET28a-PAM-K114R.
[0036] Table 1 Primers
[0037]
[0038] The PCR amplification reaction conditions are:
[0039]
[0040] PCR products were identified by agarose gel electrophoresis. Then the PCR product was purified and digested and then transformed into Escherichia coli BL21 compe...
Embodiment 2
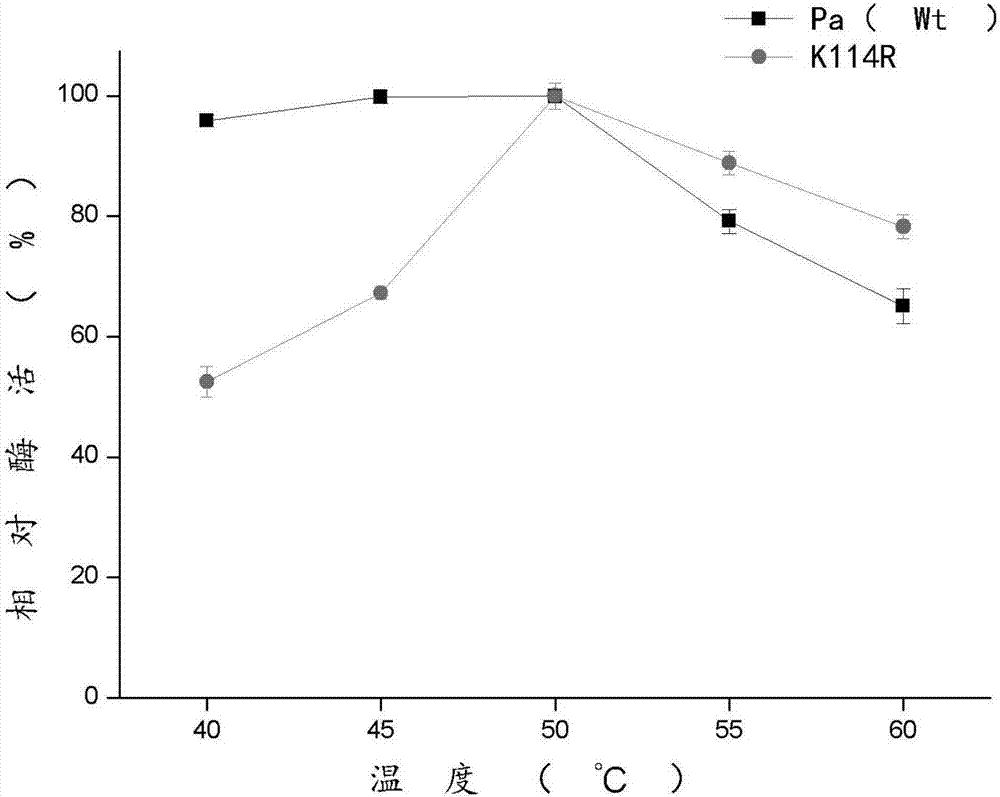
[0045] Add 100 μg of the purified mutant enzyme in Example 1 to 200 μl of the buffered reaction system, add 200 μl of the substrate L-α-phenylalanine, and react at 40°C, 45°C, 50°C, 55°C, and 60°C for 30 minutes to determine corresponding enzyme activity. The unmutated wild enzyme was used as a control, and other conditions were the same as the mutant enzyme.
[0046] Such as figure 1As shown, the relative enzymatic activity of the mutant enzyme was 51% and 63% respectively at 40°C and 45°C, and the relative enzymatic activity of the mutant enzyme was 79% and 62% at 55°C and 60°C, and that of the wild enzyme at temperature At 40°C and 45°C, the relative enzyme activities were 96% and 98%, respectively; at 55°C and 60°C, the relative enzyme activities of the mutant enzyme were 88% and 78%, respectively.
Embodiment 3
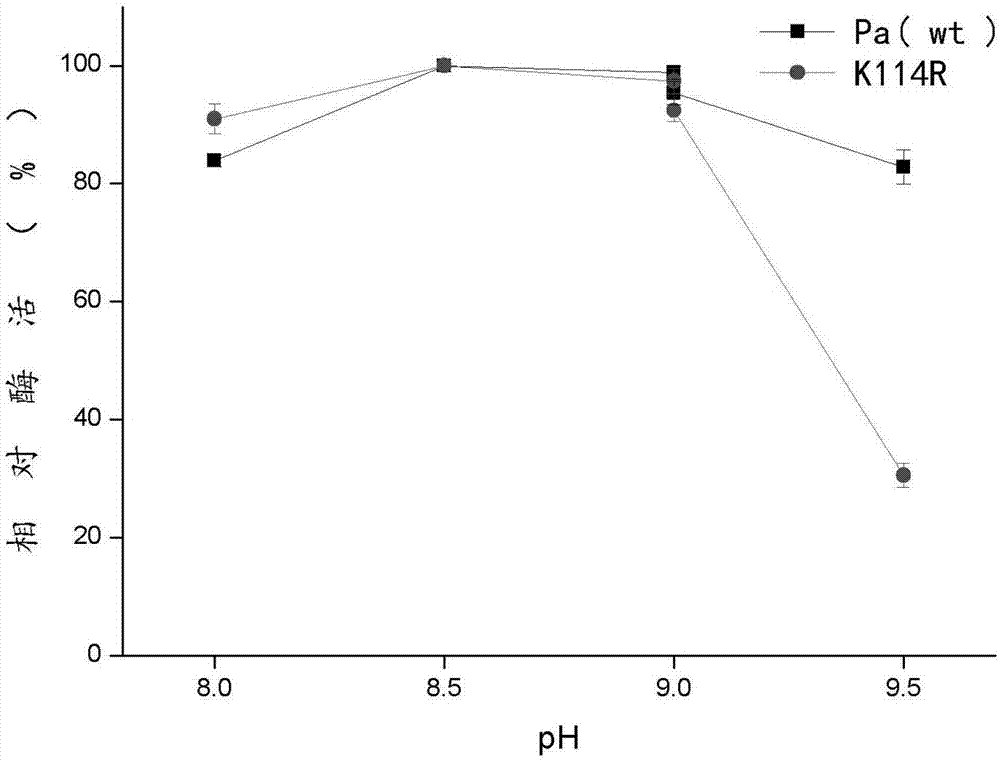
[0048] PBS buffer solutions with different pH were prepared: pH 8.0-9.0, 1 / 15mM phosphate buffer, pH=9.0-9.5, 100mM Tris-HCL buffer. The wild enzyme and the mutant enzyme were respectively reacted at 50° C. for 30 min in different pH buffer solutions, and then the enzyme activity was determined.
[0049] Such as figure 2 As shown, when pH=8.5, the enzyme activity is the highest, which is defined as 100%. When pH = 8.5, the relative activity of the wild enzyme and the mutant enzyme remained above 80% respectively, which was lower than that of the enzyme when pH = 8.5.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com