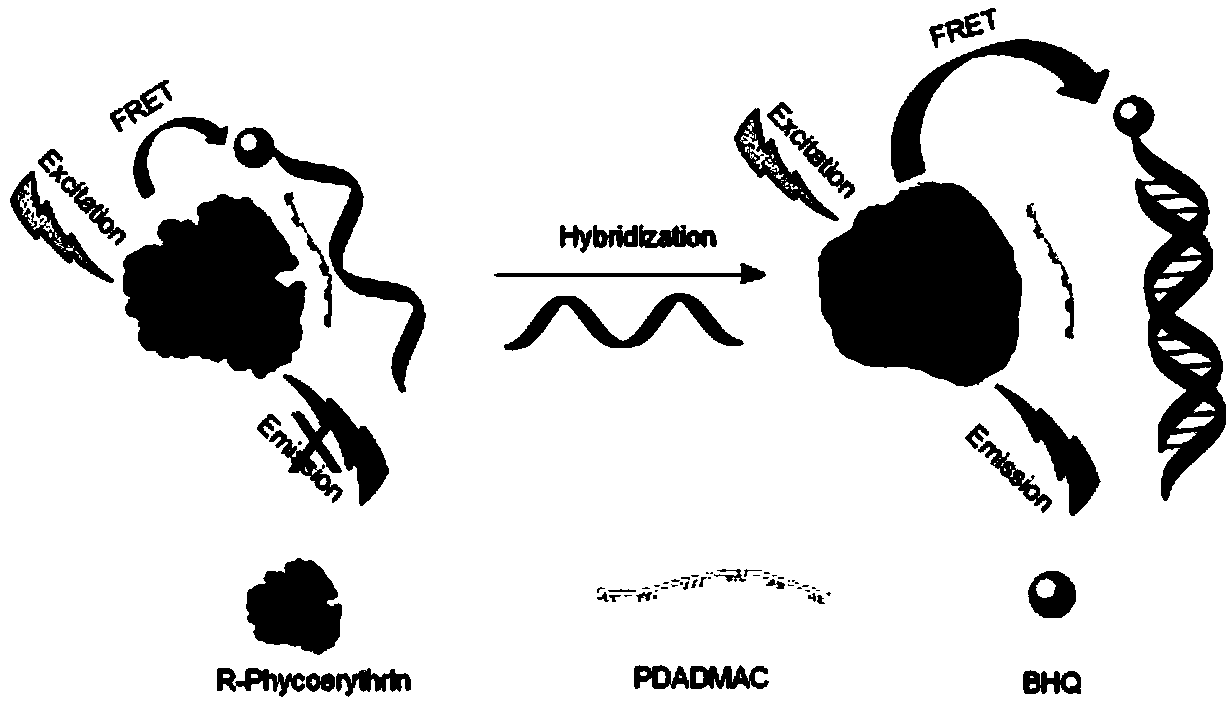
Method for detecting DNA hybridization by surface cationized R-phycoerythrin
A phycoerythrin and cationization technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, biological testing, etc., can solve the problems that DNA hybridization is not involved, and achieve short preparation time, high detection sensitivity, and detection limit low effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment one: the preparation of various solutions
[0035] (1) Preparation of 50mM PBS buffer
[0036] 29.0g Na 2 HPO 4 -12H 2 O, 2.9g NaH 2 PO 4 -2H 2 O, 5.85g NaCl was dissolved in 900ml of ultra-pure water, fixed to a 1000ml volumetric flask, and then diluted 2 times, the concentration at this time was 50mM.
[0037] (2) Preparation of phycoerythrin solution
[0038] Pipette 100 μL of phycoerythrin crystals, centrifuge at 3800 rcf at 4 °C for 15 min, discard the supernatant, and dissolve the precipitated part with 500 μL of 50 mM sodium phosphate buffer (pH7.5) for 30 min; select an ultrafiltration tube with a filter membrane of 100 kD; Filter for 15 minutes for desalting, repeat ultrafiltration for 4 times, and then dilute the concentration to 1 mg / mL with PBS buffer (pH=7.5).
[0039] (3) Preparation of Cationic Polymer PDADMAC Solution
[0040] The PDADMAC (weight<100000, 35 wt.% in H2O) solution was diluted 10 times with PBS buffer, and the concentrat...
Embodiment 2
[0045] Embodiment two: the method for DNA hybridization detection
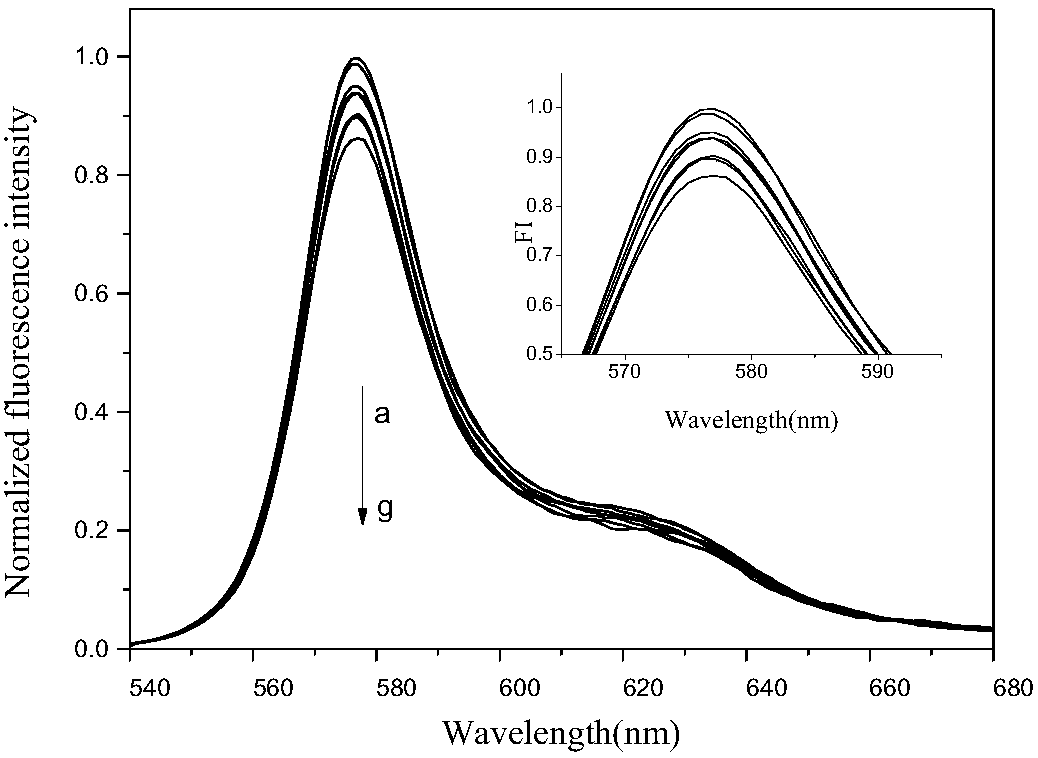
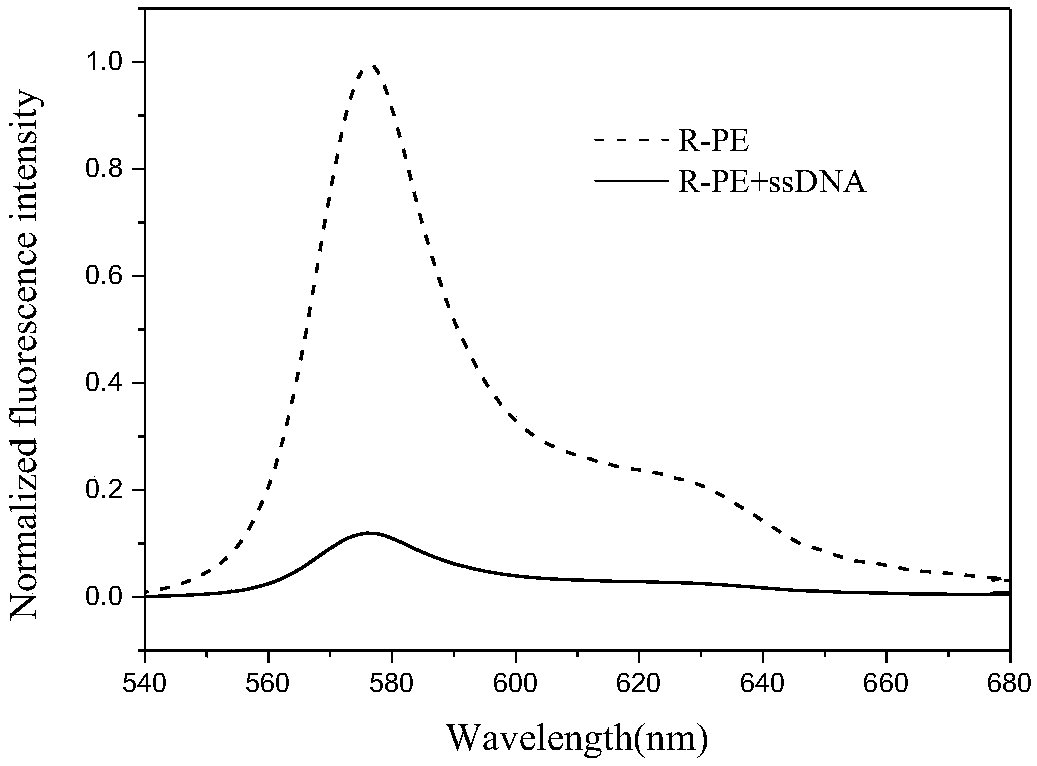
[0046] (1) After mixing R-PE and PDADMAC solution with a volume ratio of 1:10, add 5 μL of single-stranded DNA solution, then dilute to 100 μL with PBS buffer, and measure its fluorescence intensity at 575 nm two minutes later, At this time, it can be observed that the fluorescence intensity is quenched by about 88%.
[0047] (2) Mix 5 μL of single-stranded DNA solution with different concentrations of complete complementary strand solutions evenly, then add 1 μL of R-PE and 10 μL of PDADMAC solution, react for 2 minutes and dilute it to 100 μL, and measure it after two minutes The fluorescence intensity at 575nm can be observed to gradually release at this time, and finally it is basically the same as the fluorescence intensity of the protein itself.
[0048] (3) Mix 5 μL of single-stranded DNA solution with 5 μL of single-base mismatched complementary strand solution evenly, then add 1 μL of R-PE and 10 μL ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com