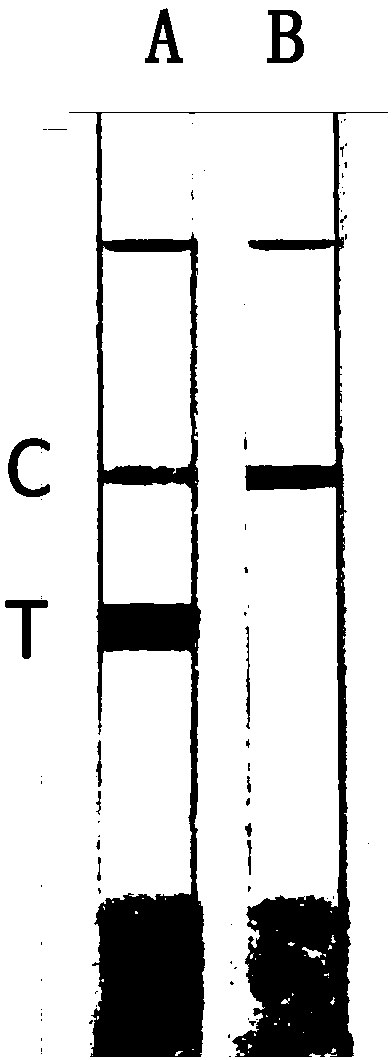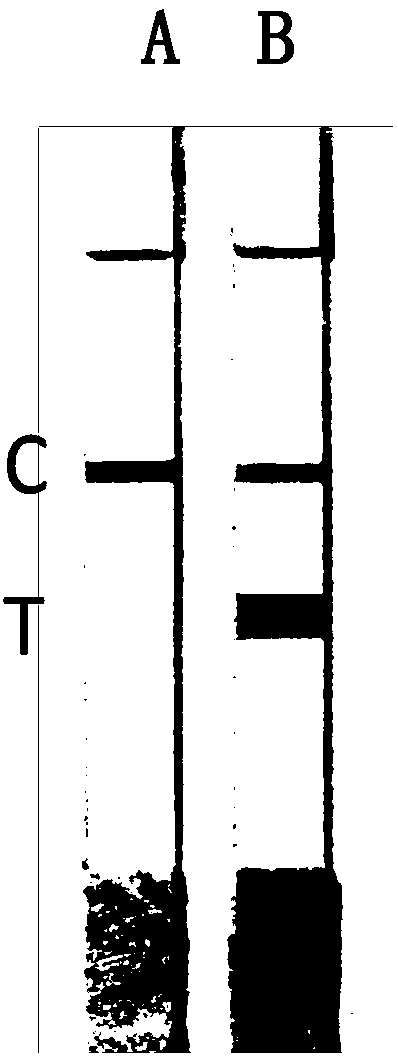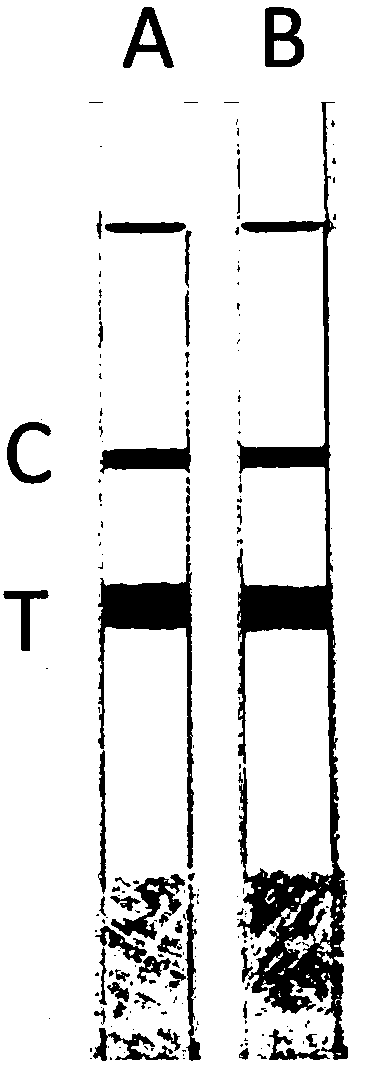Human PAH gene SNP typing rapid test paper strip detection method with characteristics of no extraction and direct amplification, and kit
A detection method and test strip technology, applied in the field of SNP detection, can solve the problems of being unsuitable for promotion and use in clinical hospitals, increasing cross-contamination of samples, and long detection time, saving the cost of DNA purification reagents, and improving detection sensitivity and accuracy. , the effect of reducing the cost of testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] The design and optimization of embodiment 1 primer
[0050] Design primers for the seven mutation sites of PAH gene: confirm the flanking sequence of the SNP site through the dbSNP database of NCBI, and design synthetic primers accordingly. For each SNP site, a wild-type specific primer was designed, whose 3' end was complementary to the wild-type base of the SNP site, and a mutant-specific primer, whose 3' end was complementary to the mutant base of the SNP site and a Common primers for identification and amplification. In order to facilitate subsequent chromatographic detection techniques, the designed primers were labeled, among which the common primers were labeled with biotin, and the specific primers were labeled with digoxigenin. In order to increase amplification specificity and detection resolution, multiple sets of primers were designed for each locus for test screening.
[0051] The optimal primers were screened according to the following objectives: (1) Hi...
Embodiment 2
[0057] Embodiment 2, the detection of whole blood sample
[0058] The whole blood samples of 10 people were taken respectively, and the mutation types were detected respectively for the seven mutation sites involved in the present invention. The specific operation steps were as follows: take a small amount of whole blood samples and add them to PCR centrifuge tubes, and draw the samples with a pipette Add 10 μL of the treatment solution into a centrifuge tube, and let it stand for 5 minutes for later use; take two PCR centrifuge tubes, A and B, and add the following substances (mutant primers are added to tube A, and wild-type primers are added to tube B):
[0059]
[0060] Put the two tubes A and B into the PCR machine, and proceed according to the optimized amplification program as follows:
[0061] 50°C UNG enzyme action for 2min, 95°C pre-denaturation for 5min, 94°C denaturation for 30s, annealing at a temperature 5°C lower than the Tm value for 30s, 65°C extension for ...
Embodiment 3
[0066] Embodiment 3: the detection of oral swab sample
[0067] Using the same sample source individuals as in Example 2, the mutation types were detected for the seven mutation sites involved in the present invention. Draw 10 μL of the sample treatment solution into the centrifuge tube, and let it stand for 5 minutes for later use; take two PCR centrifuge tubes, A and B, and add the following substances respectively:
[0068]
[0069]
[0070] Put the two tubes A and B into the PCR machine, carry out the amplification reaction and test strip detection and judgment according to the same procedure as in Example 2. The results were the same as in Example 2, and were consistent with the sequencing results. It is shown that the method is also reliable when applied to buccal swab samples.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com