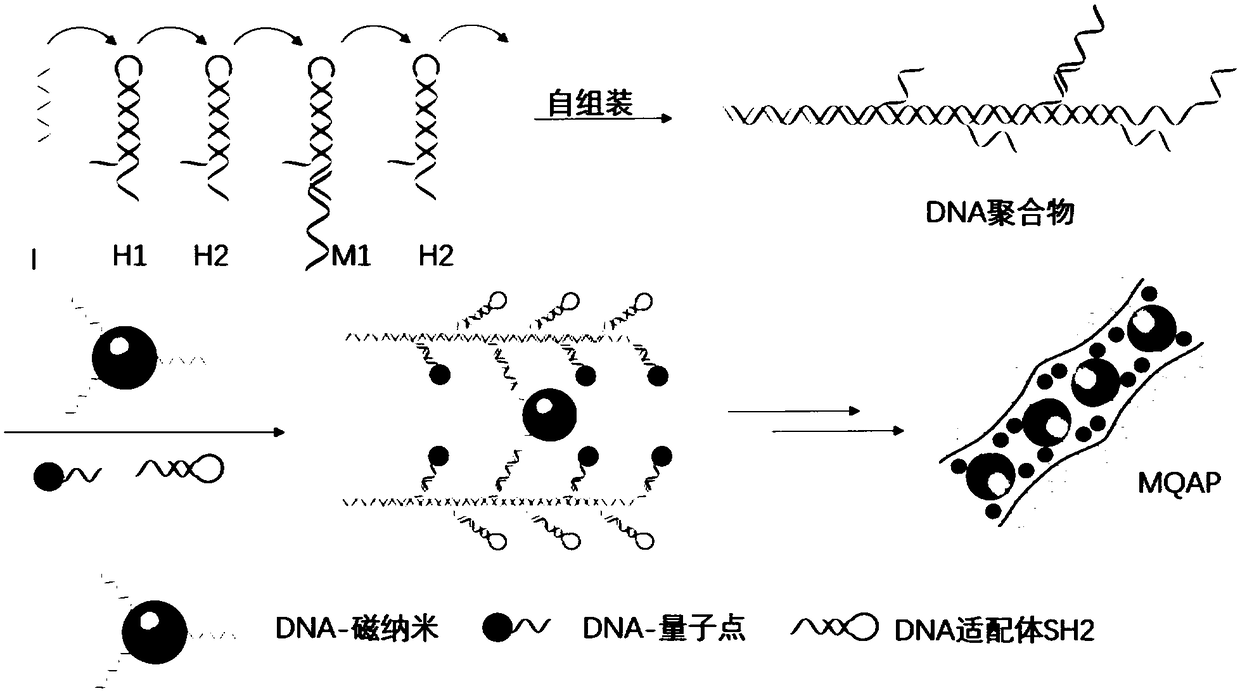
Magnetic fluorescent copolymer nano probe taking DNA (deoxyribonucleic acid) as template and application
A technology of nano-probes and copolymers, which is applied in the direction of fluorescence/phosphorescence, luminescent materials, and material analysis through optical means, which can solve the problems of high detection cost, expensive instruments and equipment, and low purity of circulating tumor cells. The effect of reducing the cost of detection and reducing the impact
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
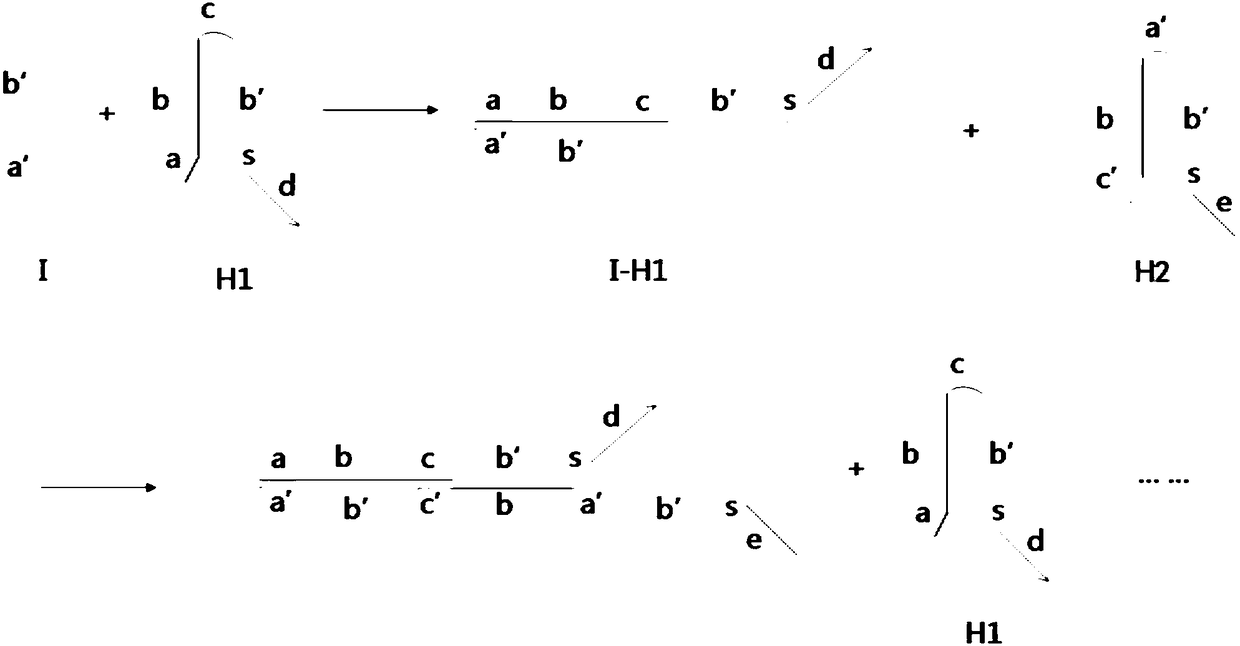
[0060] (1) Synthesis of DNA-quantum dots
[0061] In this embodiment, the C1DNA sequence used is as follows:
[0062] 5'-GGGGGGGGGGAAAAAA CGTCCGTGCTCAC -3', wherein base G has undergone thio modification.
[0063] The C2DNA sequence used is as follows:
[0064] 5'- TACGCGTCTAGGATC AAAAAAAAAAA-NH 2 -3'.
[0065] The H1 sequence used is as follows:
[0066] 5’-TTAACCCACGCCGAATCCTAGACTCAAAGTAGTCTAGGATTCGGCGTGAAAAA GTGAGCACGGACG -3', which is a hairpin structure, and the overhanging part is underlined.
[0067] The H2 sequence used was as follows:
[0068] 5'- GACGTGCAGGCTG TTTAAAGTCTAGGATTCGGCGTGGGTTAACACGCCGAATCCTAGACTACTTTG-3', which is a hairpin structure, and the overhanging part is underlined.
[0069] The promoter I sequence used is as follows:
[0070] 5'-AGTCTAGGATTCGGCGTGGGTTAA-3'.
[0071] The LH1 DNA sequence used was as follows:
[0072] 5'- GATCCTAGACGCGTA AAAAAAA CGTCCGTGCTCAC -3'.
[0073] The SH2 sequence used was as follows:
[0074] 5'- ...
Embodiment 2
[0085] Example 2 MQAP probes are used to specifically identify circulating tumor cells
[0086] (1) Culture of cell lines
[0087] The tumor cells to be detected, human acute lymphoblastic leukemia cells (CCRF-CEM) and the control cells, human B lymphocytoma cells (Ramos), were cultured. Both CCRF-CEM and Ramos cells are within 25cm 2 Cultured in the culture bottle of the culture medium, the culture medium is the RPMI 1640 culture fluid containing 1% double antibody (penicillin, streptomycin), 10% fetal bovine serum (FBS). The culture environment is 37°C, containing 5% CO 2 cell culture incubator.
[0088] (2) Specific recognition experiment of MQAP probe on CCRF-CEM cells
[0089] 200 μL of CCRF-CEM or Ramos cells (2×10 6 cells / mL) were first pre-stained with Calcein-AM (Calcein-AM, 10 μM) or 4,6-diamidino-2-phenylindole (DAPI, 10 μg / mL) in a 37°C water bath for 30 min, Add 500 μL of cell washing buffer to wash three times, and then react with 200 μL of the MQAP probe p...
Embodiment 3
[0091] Example 3 MQAP probes are used to separate and enrich CCRF-CEM cells
[0092] (1) According to the method of step (2) in Example 2, MQAP probes of different concentrations were interacted with CCRF-CEM cells, and then 100 μL (2×10 2 cells / mL) CCRF-CEM cells interacted with different concentrations of MQAP probes were added to a 24-well plate, a magnet was placed on the edge of the well plate for magnetic separation for 15 min, and then the captured tumor cells were counted by a fluorescence microscope. The cell separation device is attached Figure 7 as shown ( Figure 7 a is a schematic diagram of the separation device, Figure 7 b is the enlarged view of circle A in figure a, the arrow in the figure is the position where blood is added), and the fluorescence imaging characterization results captured by tumor cells are shown in the attached Figure 8 as shown, Figure 8 A is bright field, Figure 8 B is a quantum dot, and the arrow in the figure is a CCRF-CEM cell...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com