PCR (Polymerase Chain Reaction) amplification primer for quickly detecting klebsiella pneumonia and application thereof
A Klebsiella and amplification primer technology, applied in the field of animal bacteriology and molecular biology, can solve the problems of long time consumption and insensitive detection, and achieve the effects of less time consumption, high sensitivity and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Establishment of a PCR method for rapid detection of Klebsiella pneumoniae
[0038] 1. Preparation of materials
[0039] Klebsiella pneumoniae, Cryptobacterium cryptica, Mannella hemolyticus, Pasteurella, Mycoplasma bovis, bovine parainfluenza virus type 3, bovine infectious rhinotracheitis virus and Escherichia coli were isolated, identified and preserved by Guangxi Veterinary Research Institute. Samples were obtained from veterinary clinics. 10×PCR Buffer, dNTPs, ES-Taq DNA polymerase, DNA / RNA extraction kit, bacterial genome DNA extraction kit were purchased from Kangwei Century Biotechnology Co., Ltd.
[0040] 2. Design and synthesis of PCR primers
[0041] According to the homology comparative analysis of the phoE gene sequence of Klebsiella pneumoniae in GenBank, the conserved sequence region was selected as the amplification region, and specific amplification primers were designed by using Oligo 7.0 primer design software and BLAST software program. ...
Embodiment 2
[0072] Example 2 Rapidly detects the annealing temperature test of Klebsiella pneumoniae PCR method
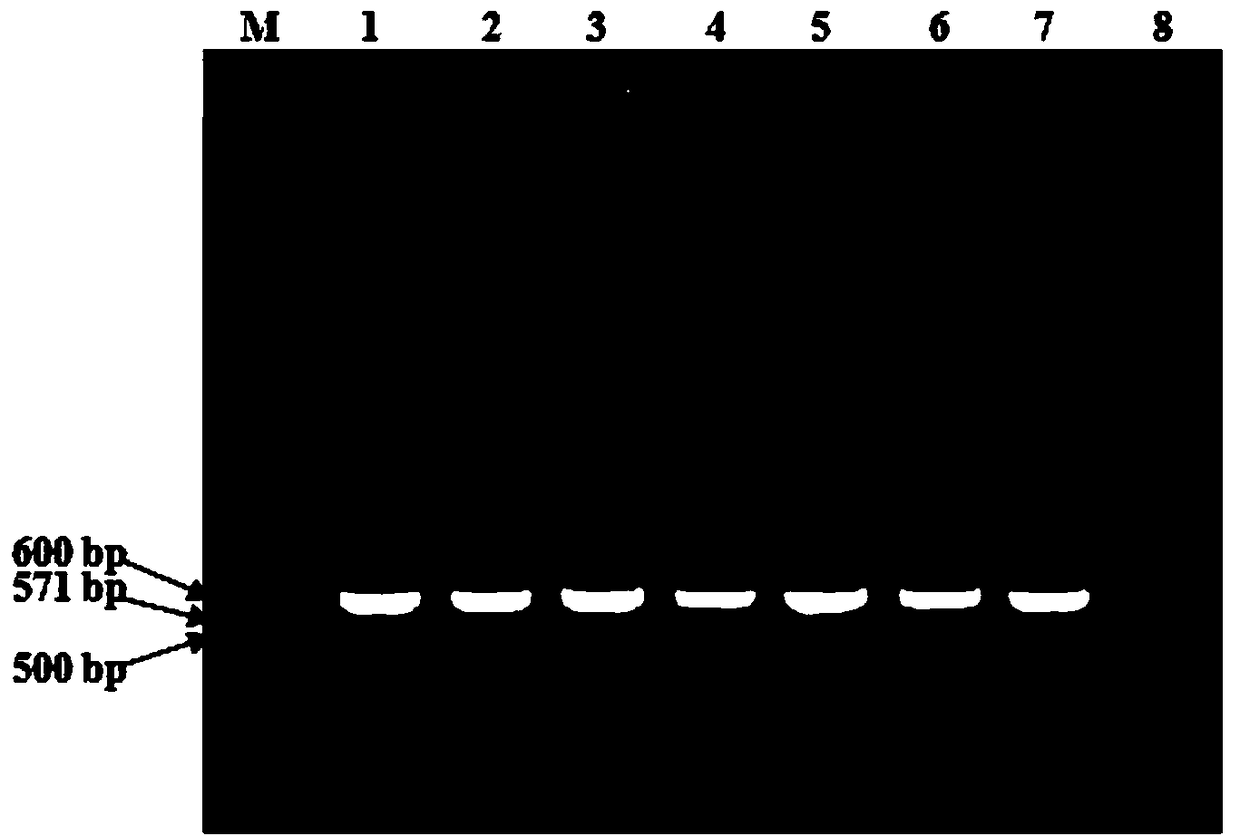
[0073] Perform PCR amplification at annealing temperatures of 50 °C, 52 °C, 54 °C, 56 °C, 58 °C, 60 °C, and 62 °C to determine the optimal annealing temperature. The results showed that the designed primers had a large tolerance to annealing temperature, and could amplify well under the reaction programs with annealing temperatures of 50 ℃, 52 ℃, 54 ℃, 56 ℃, 58 ℃, 60 ℃ and 62 ℃. Add a single destination strip ( figure 1 ). To this end, the reaction program used in this PCR method was: 95°C for 5 min; followed by 35 cycles of 95°C for 35 s, 58°C for 40 s, and 72°C for 45 s; and finally 72°C for 10 min.
Embodiment 3
[0074] Embodiment 3 detects the specific detection result of Klebsiella pneumoniae PCR method
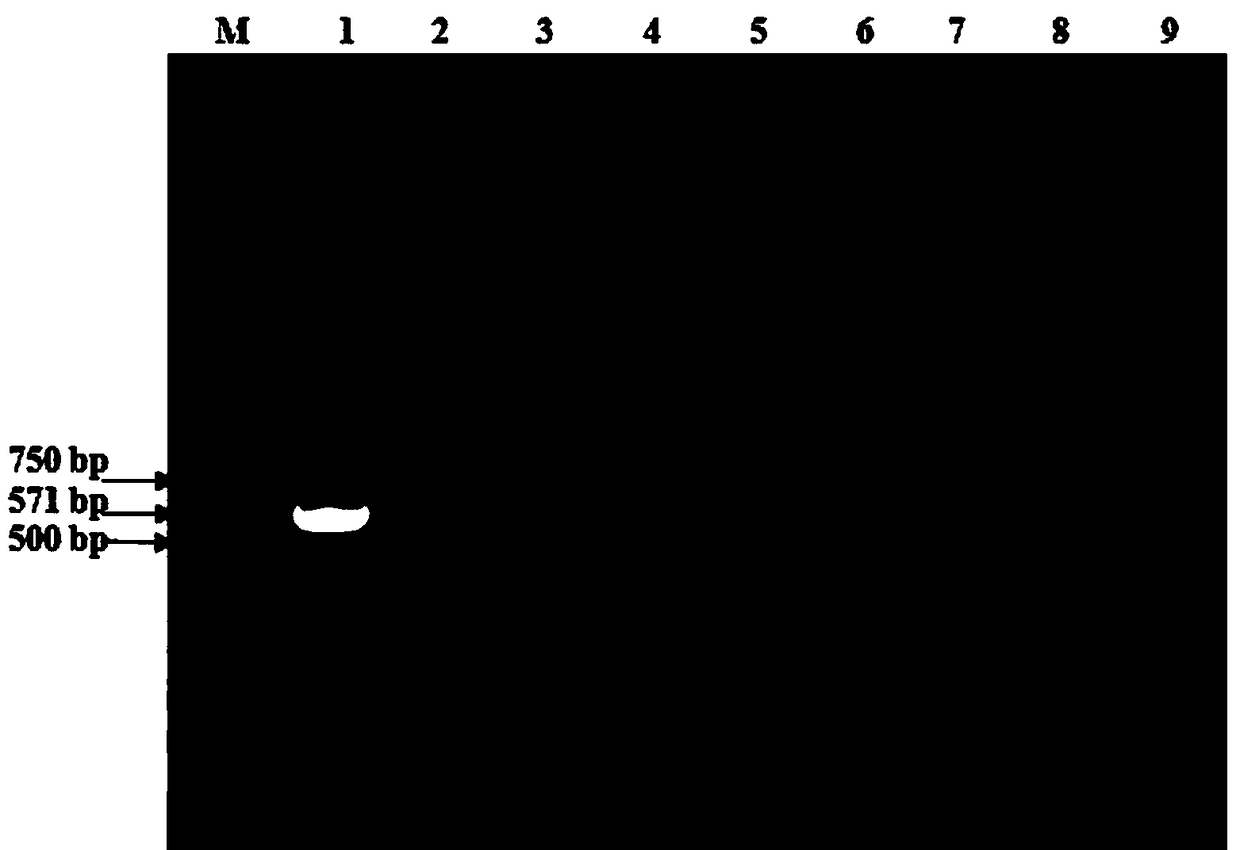
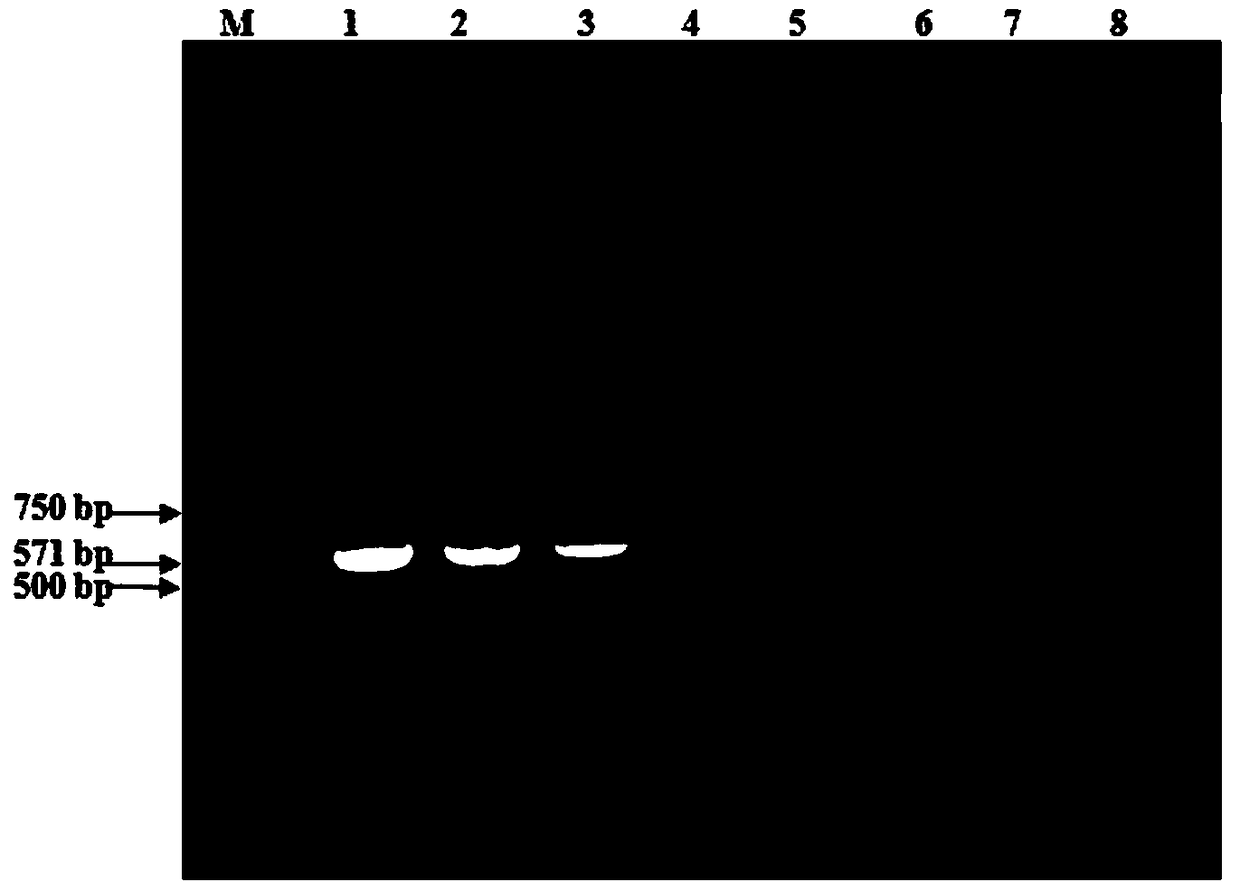
[0075] Genomic DNA / RNA (RNA for reverse reaction) was extracted from Klebsiella pneumoniae, Cryptobacterium, Mannella hemolyticus, Pasteurella, Mycoplasma bovis, Bovine parainfluenza virus type 3, Bovine infectious rhinotracheitis virus and Escherichia coli. Transcribed into cDNA), using the optimized reaction system and reaction program for PCR amplification, testing the specificity of the detection method of the present invention, the results showed that only the Klebsiella pneumoniae sample amplified the target fragment band, for Positive results, no amplified bands were detected in the reaction tubes of the 7 strain control strains and the water control reaction tube, which was a negative result ( figure 2 ), indicating that the method has good specificity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com