Method of excavating mass spectrum data and screening specific marker of red ginseng
A technology of mass spectrometry data and screening methods, which is applied in the field of rapid discovery of red ginseng biomarkers, mining and screening of specific markers, can solve the problems of storage and processing indefinitely, exact definition fuzzy, etc., and achieve weakening ion suppression , high accuracy, and the effect of improving analytical throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] 1. Extraction of red ginseng and ginseng compounds:
[0033] Solvent extraction: crush 50 red ginseng from different sources (different pharmacies) and 50 white ginseng medicinal materials from different sources (different pharmacies), take 100 mg respectively, put them in 2 ml centrifuge tubes, add 0.5 ml volume concentration to be 10 % of acetonitrile aqueous solution is the extract, mix well; ultrasonic (1130W, 37 kHz) for 15 minutes, separate, collect the solvent, repeat the extraction 2 times, combine the extracts to obtain extract 1; add 0.5ml of dregs to make the concentration 80% Acetonitrile water, mixed evenly, ultrasonic for 15 minutes, separated, collected solvent, repeated extraction 2 times, combined extracts to obtain extract 2; add 8 μL DMSO to extracts 1 and 2 respectively, and concentrate in vacuo (remove acetonitrile , water), to obtain Concentrate 1 and Concentrate 2 respectively; Dissolve Concentrate 1 and Concentrate 2 in 200 μL of 10% acetonitrile...
Embodiment 2
[0053] 1. Extraction of red ginseng and ginseng compounds:
[0054] Solvent extraction: crush 50 red ginseng from different sources (different pharmacies) and 50 white ginseng medicinal materials from different sources (different pharmacies), take 100 mg respectively, put them in 2ml centrifuge tubes, add 0.4ml volume concentration to be 20 % of acetonitrile aqueous solution is the extract, mix well; ultrasonic (1130W, 37 kHz) for 20 minutes, separate, collect the solvent, repeat the extraction 3 times, combine the extracts to obtain the extract 1; add 0.4ml of dregs and the concentration is 90% Acetonitrile and water, mixed evenly, ultrasonicated for 20 minutes, separated, collected the solvent, repeated extraction 3 times, combined the extracts to obtain the extract 2; respectively added 10 μL dimethyl sulfoxide to the extracts 1 and 2, concentrated in vacuo (removed acetonitrile , water), to obtain concentrate 1 and concentrate 2 respectively; concentrate 1 and concentrate ...
example 2
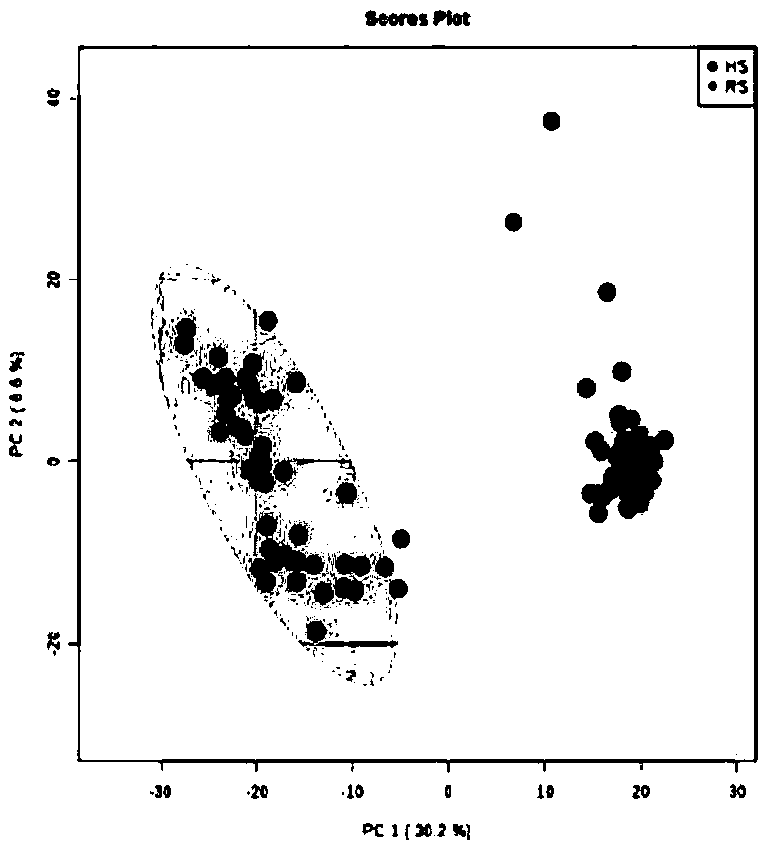
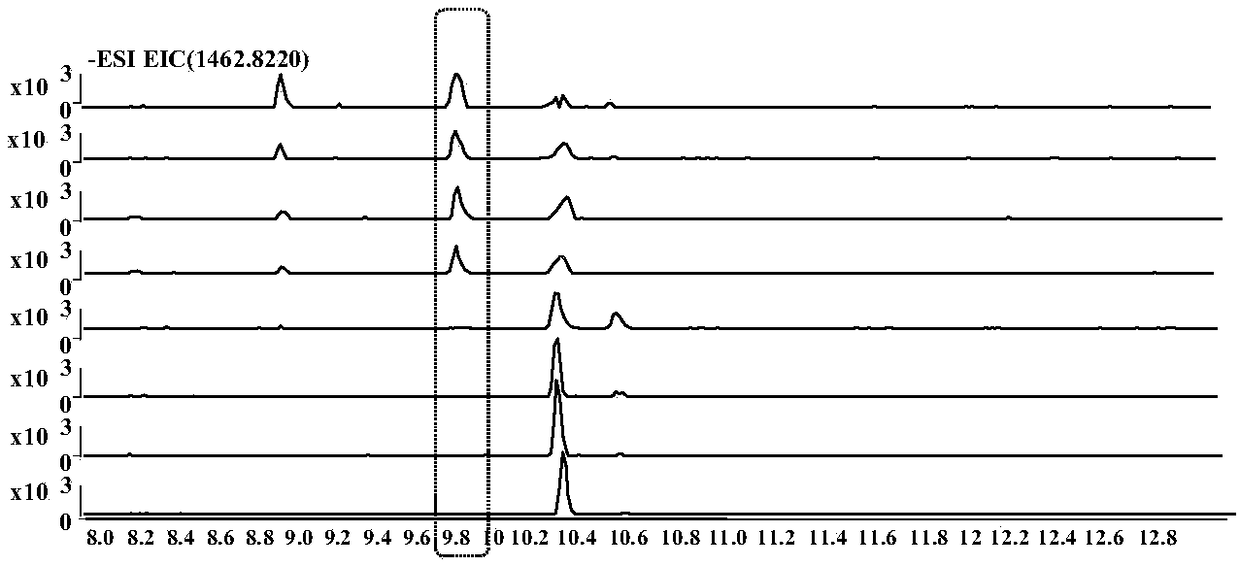
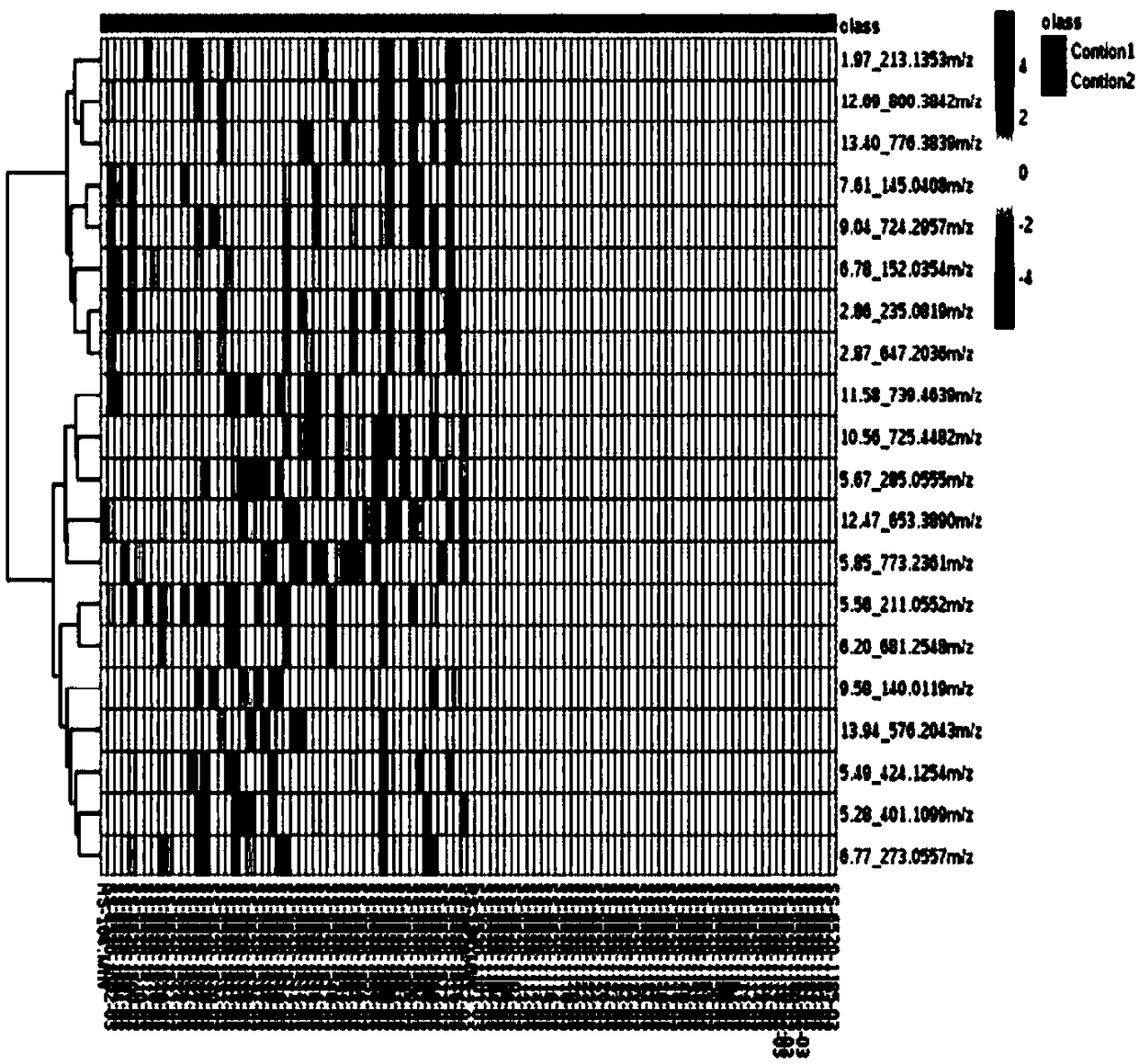
[0069] Example 2 result and table 1 in example 1, Picture 1-1 , Figure 1-2 , figure 2 , Figure 3-1 , Figure 3-2 The results are consistent, and the prediction accuracy rate is 100%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com