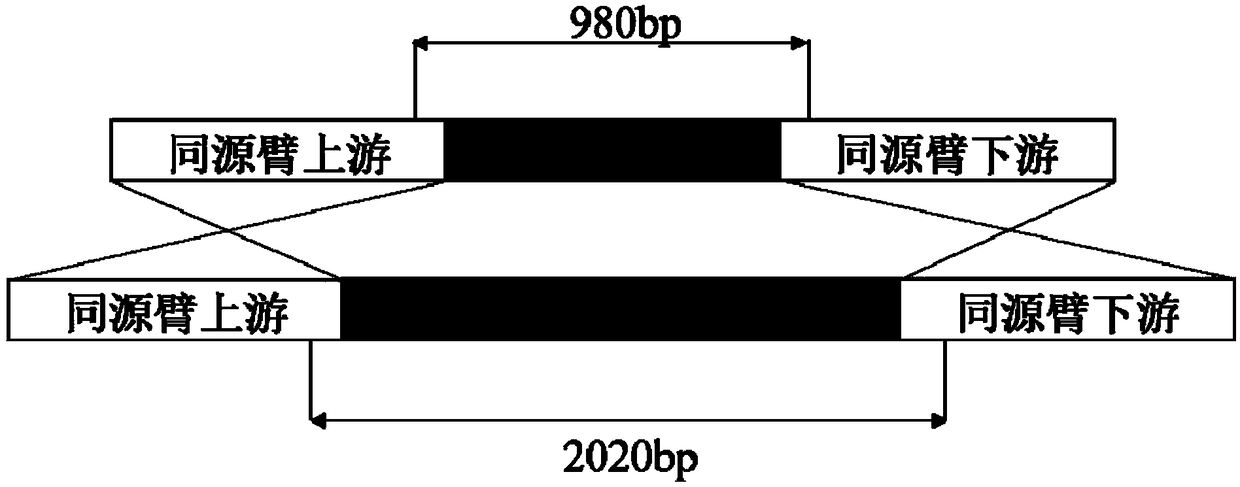
Preparation method of thermomyces PCR template
A fungal and template technology, applied in microorganism-based methods, biochemical equipment and methods, and microbial assay/inspection, etc., can solve the problems of unfavorable high-throughput screening of transformants, cumbersome operations, and large amount of bacterial cells.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Construction of knockout plasmids
[0042] Inoculate Dupont thermophiles NRRL 2155 wild bacteria from slant medium onto PDA medium, place them in a 45°C incubator and cultivate them for 7 days, then scrape about 100 mg of mycelia, and use the CTAB method to extract genomic PCR templates. The steps are as follows:
[0043] (1) Add 10mL of CTAB separation buffer (CTAB 4g / L, NaCl 16.364g / L, 1M Tris-HCl 20ml) into a 50mL centrifuge tube and preheat in a 60°C water bath.
[0044] (2) Weigh 100 mg of mycelium, place it in a pre-cooled mortar, pour liquid nitrogen into it, and grind the mycelia as soon as possible.
[0045] (3) Add the ground powder directly into the preheated CTAB separation buffer, and gently swirl to make it evenly mixed.
[0046] (4) The sample was incubated at 65°C for 30 minutes.
[0047] (5) Add an equal volume (10 mL) of chloroform-isoamyl alcohol, and gently invert to mix.
[0048] (6) Centrifuge at 4000 rpm for 10 minutes at room temperature.
[...
Embodiment 2
[0058] Preparation, transformation and validation of protoplasts from Dupontella thermophilica NRRL 2155
[0059] Preparation of TE buffer: 10mM Tris-HCl and 0.1mM EDTA, pH 7.5.
[0060] (1) Inoculate the mycelium block of Dupont thermophilic bacteria NRRL 2155 in the center of the PDA medium, and place it in an incubator at 45°C for 7 days. Use 1mL pipette tip and sterile water (add 0.05% Tween 20) to scrape the culture from the above plate, filter with four layers of lens paper, and divide the liquid containing spores into 1.5mL centrifuge tubes, 10000rpm , centrifuge at room temperature for 5 min to enrich the spores, discard the supernatant, enrich to 1×10 8 cells / mL and washed twice with sterile water.
[0061] (2) Transfer 200 μL of the spore suspension to 100 mL of YPS liquid medium, culture at 45° C. and shake at 180 rpm for 20 h.
[0062] (3) Pour the mycelium cultivated in step (2) into a sterile funnel (containing four layers of lens-cleaning paper) and filter to...
Embodiment 3
[0081] The thermophilic Dupont bacteria NRRL 2155 wild bacteria were inoculated on the PDA medium from the slant medium, and placed in a 45°C incubator for 5 days.
[0082] The PCR template preparation was the same as step (17) in Example 2. The difference is that the mycelium selected is the hyphae of the wild fungus cultured in this example; and the formula of the TE buffer is: 8mM Tris-HCl and 0.05mM EDTA, pH 7.
[0083]The PCR verification operation is the same as step (18) in Example 2. The difference is that the primers used in PCR are ER5F (SEQ ID No.1) / ER5R (SEQ ID No.2), and the PCR template is the template prepared in this example.
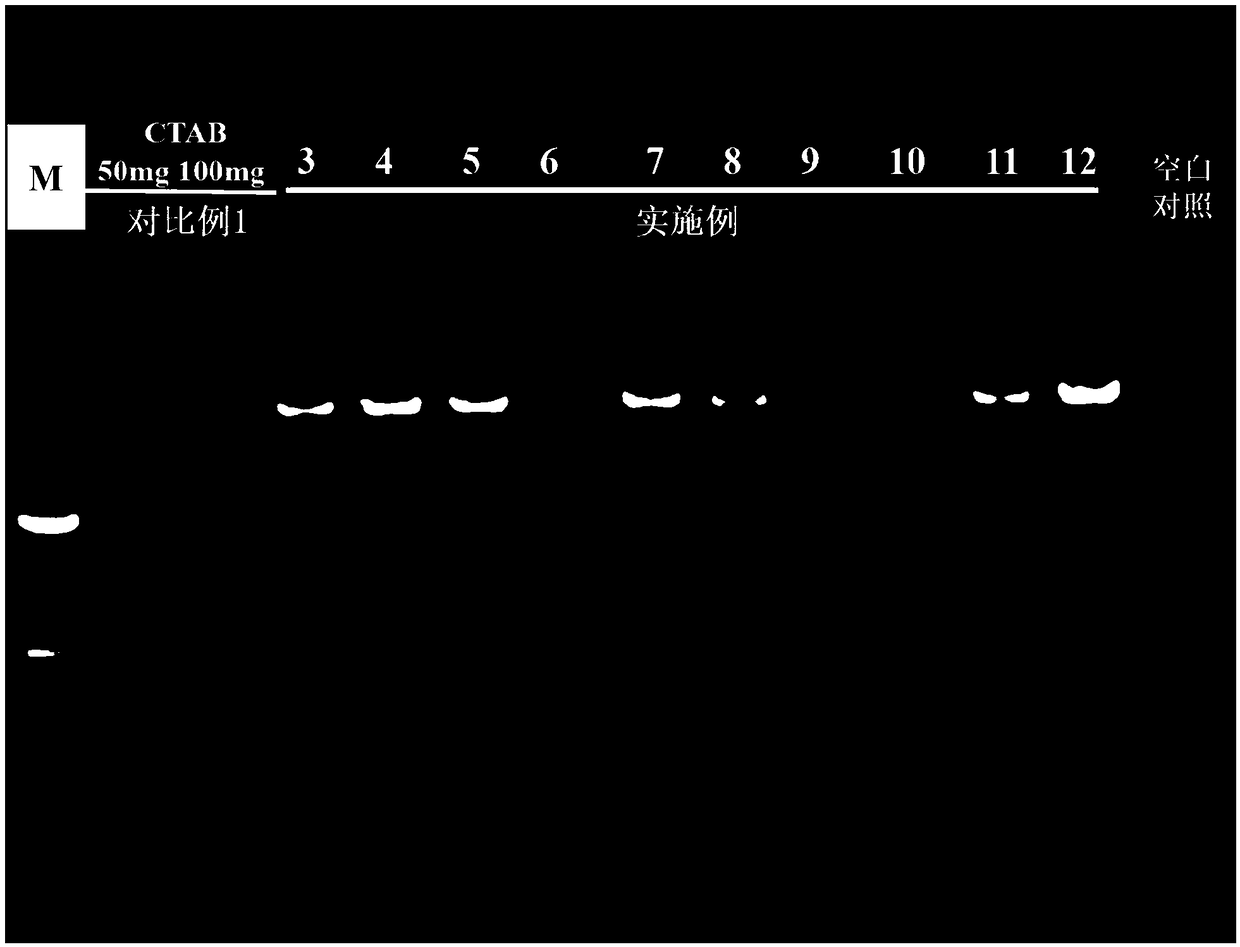
[0084] see results image 3 Lane 3 in the figure, it can be judged from the figure that a clear target band can be effectively amplified, but there are fuzzy non-specific bands below the target band. Therefore, although the quality of the lysed bacterial cell suspension obtained under the conditions of this example is not the best, it...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com