Preparation method of capture probe of large-fragment genome as well as kit
A technology for capturing probes and genomes, applied in the field of preparation and processing of biological samples, can solve the problems of unsuitable capture of a large number of regions, lower probe binding efficiency, deviation, etc., and achieve the effect of improving binding efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
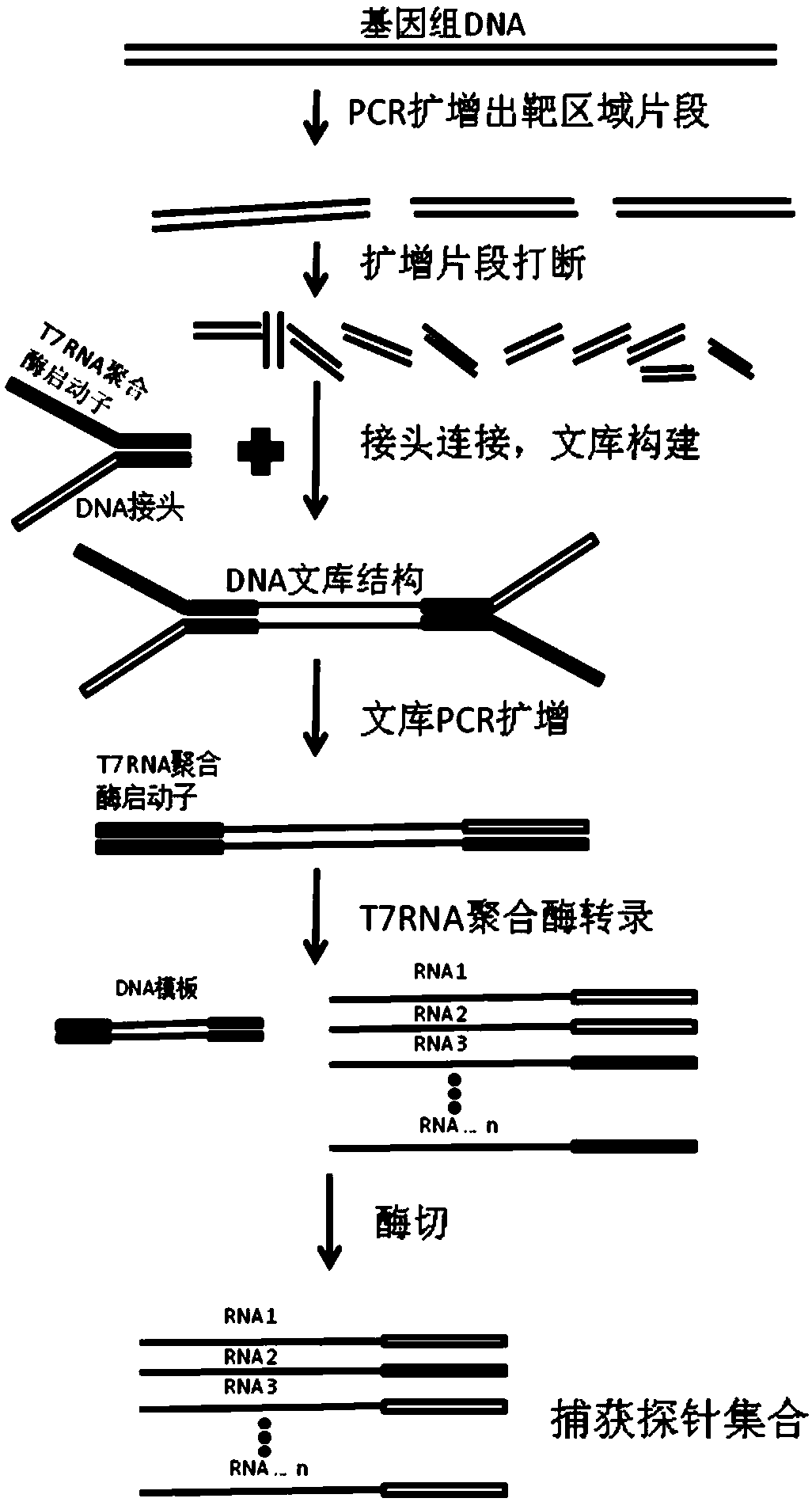
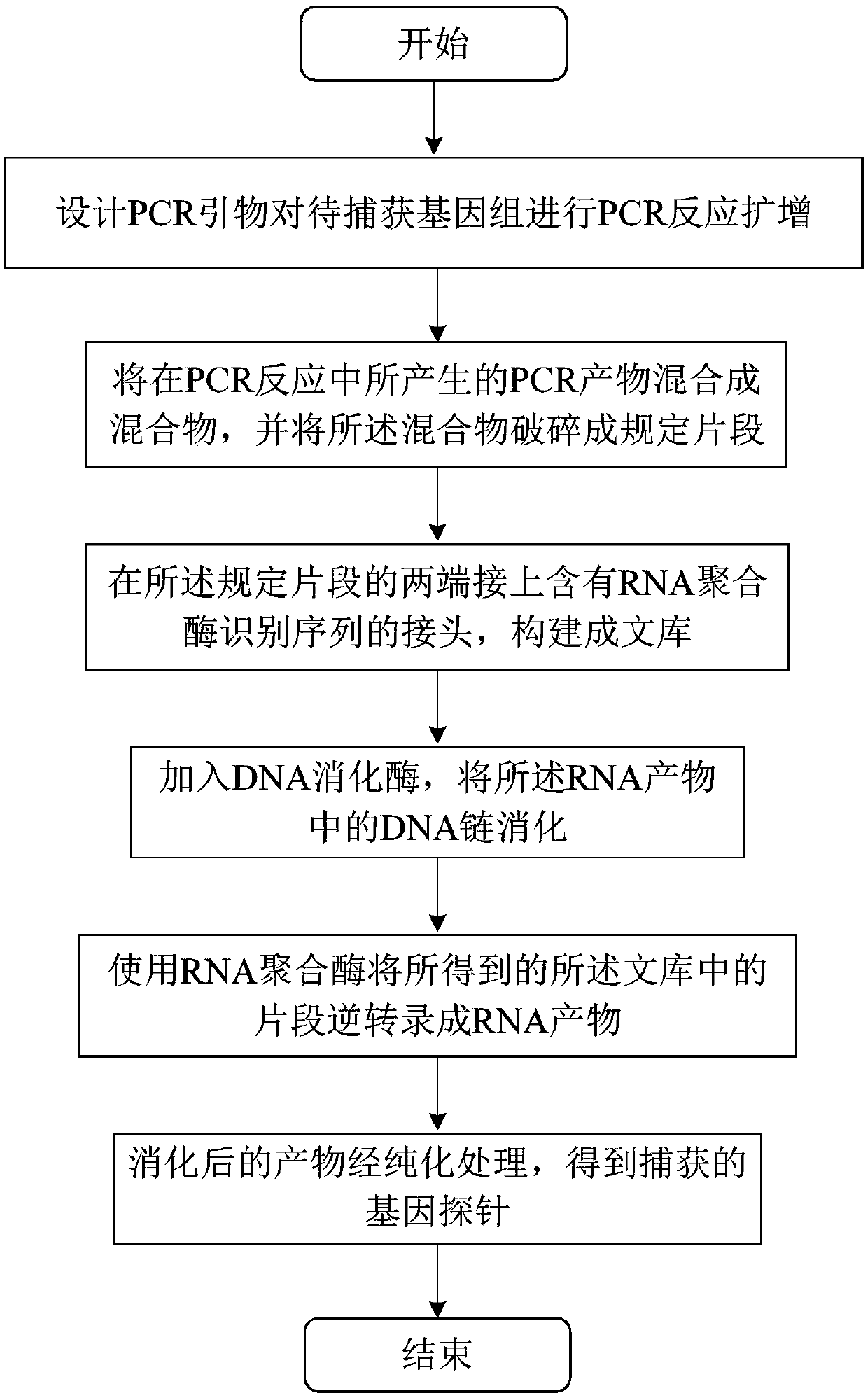
[0023] The preparation method of the large-fragment region genome capture probe involved in this embodiment includes: designing PCR primers to perform PCR reaction amplification of the genome to be captured (step S10); mixing the PCR products generated in the PCR reaction into a mixture, and Fragmentation of the mixture into prescribed fragments (step S20); linkers containing RNA polymerase-recognizable promoter sequences are connected to both ends of the prescribed fragments to construct a library (step S30); The fragments are reverse transcribed into RNA products (step S40); DNA digestion enzymes are added to digest the DNA strands in the RNA products (step S50); and the digested products are purified to obtain captured gene probes (step S60).
[0024] In the preparation method involved in this embodiment, the amplified product of the gene in the large fragment region to be captured is broken into predetermined fragments, and a gene probe (RNA probe) of suitable length is obt...
Embodiment 1
[0039] This example is carried out by using self-prepared reagents for library construction, and the post-enzyme digestion treatment method is USER digestion.
[0040]
[0041] 1. PCR amplification of target region fragments
[0042] PCR primers are commissioned to be synthesized by commercial companies, limited to the ability to amplify 200bp-20kb length fragments on the genome. For each pair of PCR primers, a 0.2ml PCR tube was used for PCR reaction, and the reaction system was shown in Table 1.
[0043] Table 1
[0044]
volume
Phusion High-Fidelity PCR mix
15μl
PCR Forward Primer F (SEQ ID NO.1, 20uM)
1μl
PCR reverse primer R (SEQ ID NO.2, 20uM)
1μl
10-200ng
Supplement H 2 O to
30μl
[0045] Perform the following reaction program in the PCR instrument (see Table 2).
[0046] Table 2
[0047]
step
temperature
time
activation
1
95℃
3min
tra...
Embodiment 2
[0129] In this example, commercial kits were used for library construction, and the post-digestion treatment was DNaseI digestion.
[0130]
[0131] 1. PCR amplification of target region fragments
[0132] The PCR product is commissioned to be synthesized by a commercial company, limited to the ability to amplify a 200bp-20kb length fragment on the genome.
[0133] Using a 0.2ml PCR tube, perform the following PCR reactions for each pair of PCRs.
[0134] Table 16
[0135]
[0136] PCR reaction conditions:
[0137] Table 17
[0138]
[0139] After the PCR is completed, all products are subjected to DNA purification. The purification steps can be carried out according to the operation steps of Sangon’s PCR Purification Kit (Cat. No.: B518141), or purification kits with similar functions from other companies can also be used.
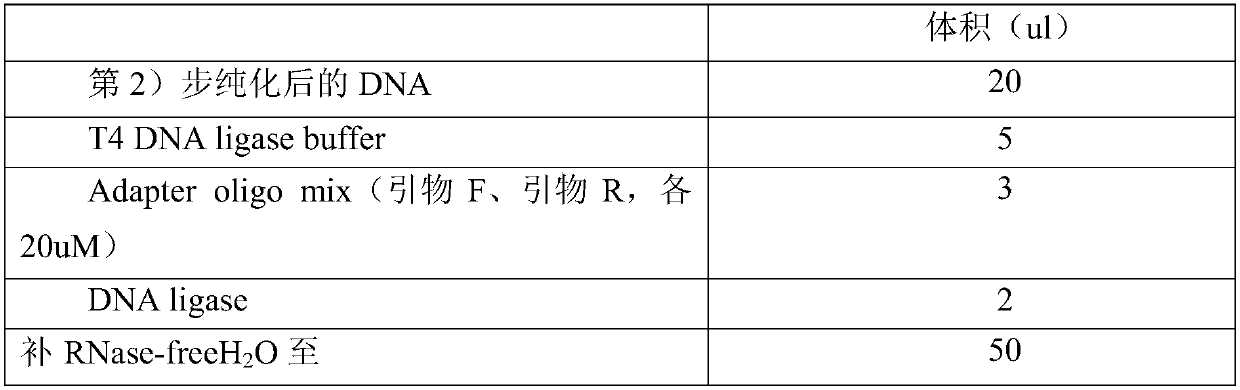
[0140] 2. Mix the above PCR products with equal molecular weights; after mixing, take 500ng and cut the PCR products to about 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com