Duplex nanometer PCR detection method for identifying CCoVI and CCoVII
A detection method and nanotechnology, applied in the field of genetic engineering, can solve problems such as unsuitable canine viruses, achieve good sensitivity, high sensitivity and specificity, and avoid the effect of non-specific amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: CCoVI and CCoVII RT-PCR amplification of target genes
[0039] 1. Primer design:
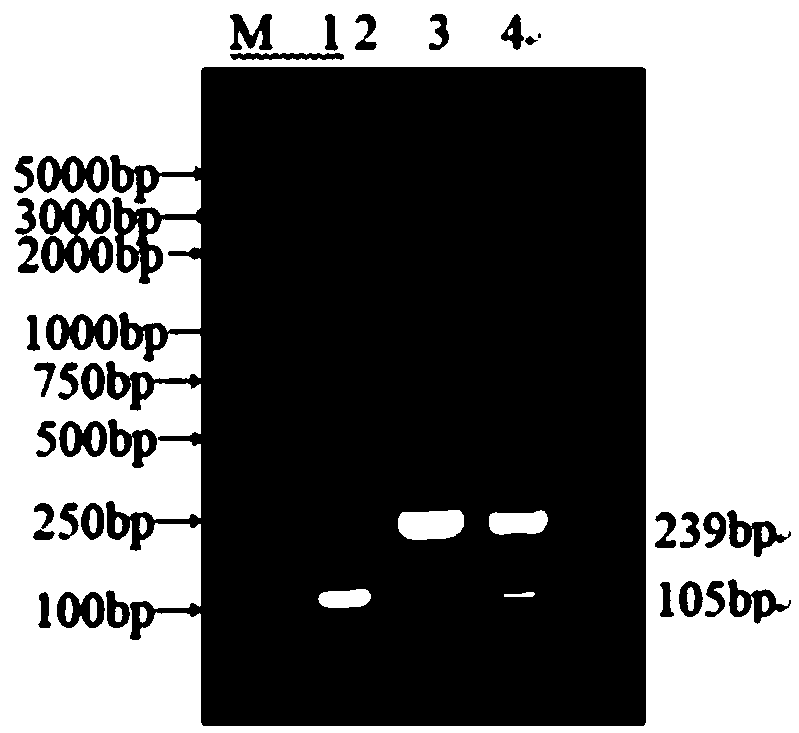
[0040] The primers CCoVII-F and CCoVII-R of the present invention are used to amplify the specific sequence of CCoVII, and the size of the amplified fragment is 239bp; the primers CCoVII-F and CCoVII-R are used to amplify the specific sequence of CCoVII, and the size of the amplified fragment is 105bp. Primers were synthesized by Beijing Huada Biotechnology Co., Ltd. The primer sequences are detailed below:
[0041] CCoVI-F: 5'-GTGCTTCCTCTTGAAGGTACA-3' (SEQ ID NO.1);
[0042] CCoVI-R: 5'-TCTGTTGAGTAATCACCAGCT-3' (SEQ ID NO.2);
[0043] CCoVII-F: 5'-TAGTGCATTAGGAAGAAGCT-3' (SEQ ID NO.3);
[0044] CCoVII-R: 5'-GCAATTTTGAACCCTTC-3' (SEQ ID NO. 4).
[0045] The nucleotide sequences of specific target fragments obtained after amplification are shown in SEQ ID NO.5 (CCoVI) and SEQ ID NO.6 (CCoVII).
[0046] 2. Establishment of common RT-PCR detection methods for CCoVI and CCoV...
Embodiment 2
[0049] Embodiment 2: The establishment of CCoVI and CCoVII nanometer RT-PCR (nanoRT-PCR) detection method
[0050] 1. Optimization of annealing temperature for CCoVI and CCoVII nanoRT-PCR:
[0051] Using the recombinant plasmids pMD-CCoVI and pMD-CCoVII as templates, the annealing temperature was set at 46°C to 53°C using a temperature gradient PCR instrument, and the annealing temperature was optimized by increasing by 1°C. The reaction system was as follows: Taq DNA polymerase (5U / μL) (GRED, Shandong, China), 0.5 μL; 2×nano PCR Buffer (GRED, Shandong, China), 10 μL; CCoVI-F, CCoVI-R, CCoVII-F and 1 μL each for CCoVII-R; 1.0 μL each for pMD-CCoVII and pMD-CCoVII; add ddH2O to 20 μL. The reaction conditions were as follows: 95°C, 3min; 35 cycles: 95°C, 30s, 46°C-53°C (1°C in order), 30s, 72°C, 30s; 72°C, 10min.
[0052] CCoVI and CCoVⅡ nanoRT-PCR was optimized by annealing temperature (46℃~53℃), and the results of agarose gel electrophoresis are shown in figure 2 . It can...
Embodiment 3
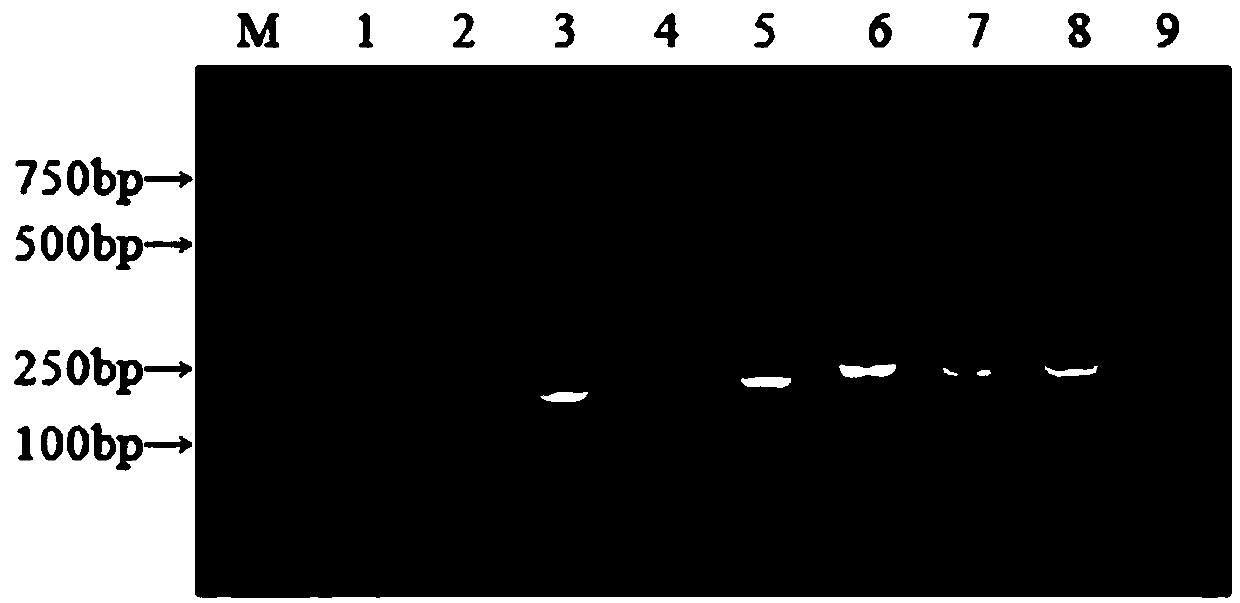
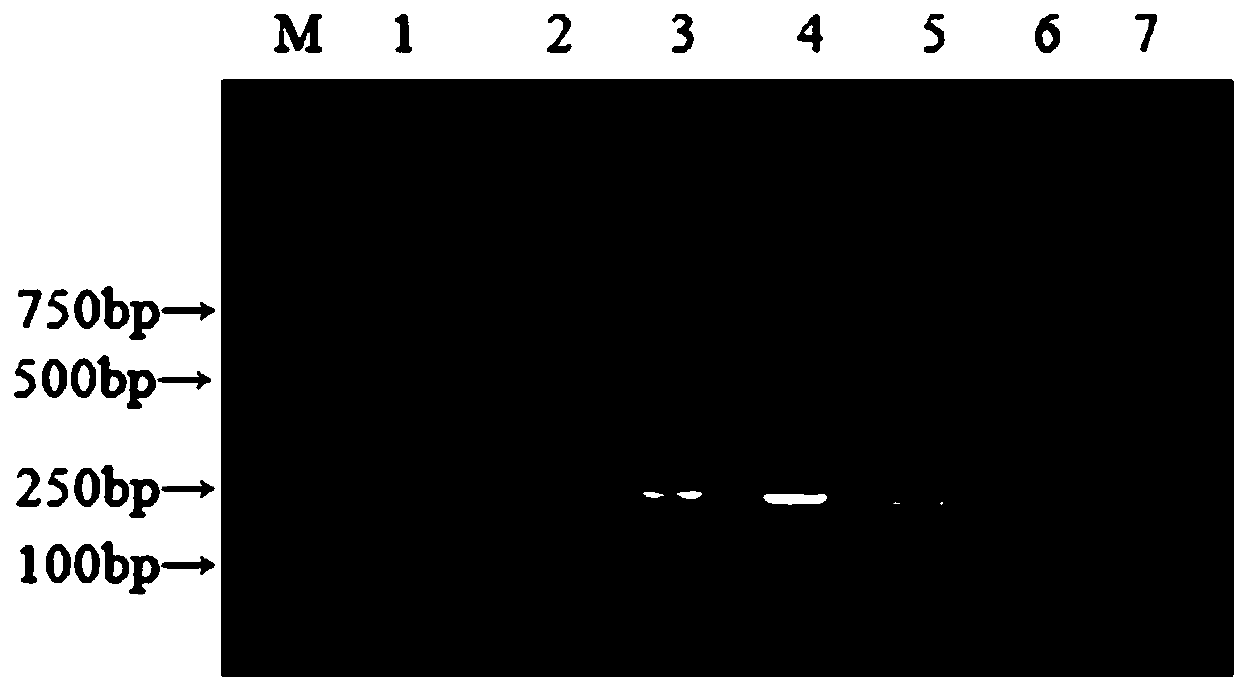
[0064] Example 3: Detection and verification of clinical samples
[0065] 1. Collection and processing of clinical samples:
[0066] A total of 60 dog feces samples were obtained from an animal hospital in Beijing. A small amount of dog feces was taken and diluted 1:4 with 0.01mol / LPH7.6PBS. After centrifugation at 12000r / min for 5min, the supernatant was taken and applied with Axyprep Body FluidViral DNA Genomic RNA was extracted using the / RNA Miniprep kit. Using this RNA as a template, cDNA was synthesized according to the instructions of the M-MLV reverse transcription kit, and the cDNA of the obtained sample was stored at -20°C until testing.
[0067] 2. Detection and verification:
[0068] CCoVI and CCoVII were detected on cDNA of 60 clinical samples by using nanoRT-PCR established in the present invention. The test results showed that the positive rate of CCoVII was 11.67%, the positive rate of CCoVII was 48.33%, and the mixed infection rate of the two was 8.33% (see T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com