Engineering bacteria and application in coproduction of tanshinol and alanine
A technology of amino acid and transaminase, applied in the field of bioengineering, can solve problems such as high cost and complicated operation, and achieve the effects of simple production process, easy-to-obtain raw materials, and strong optical specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
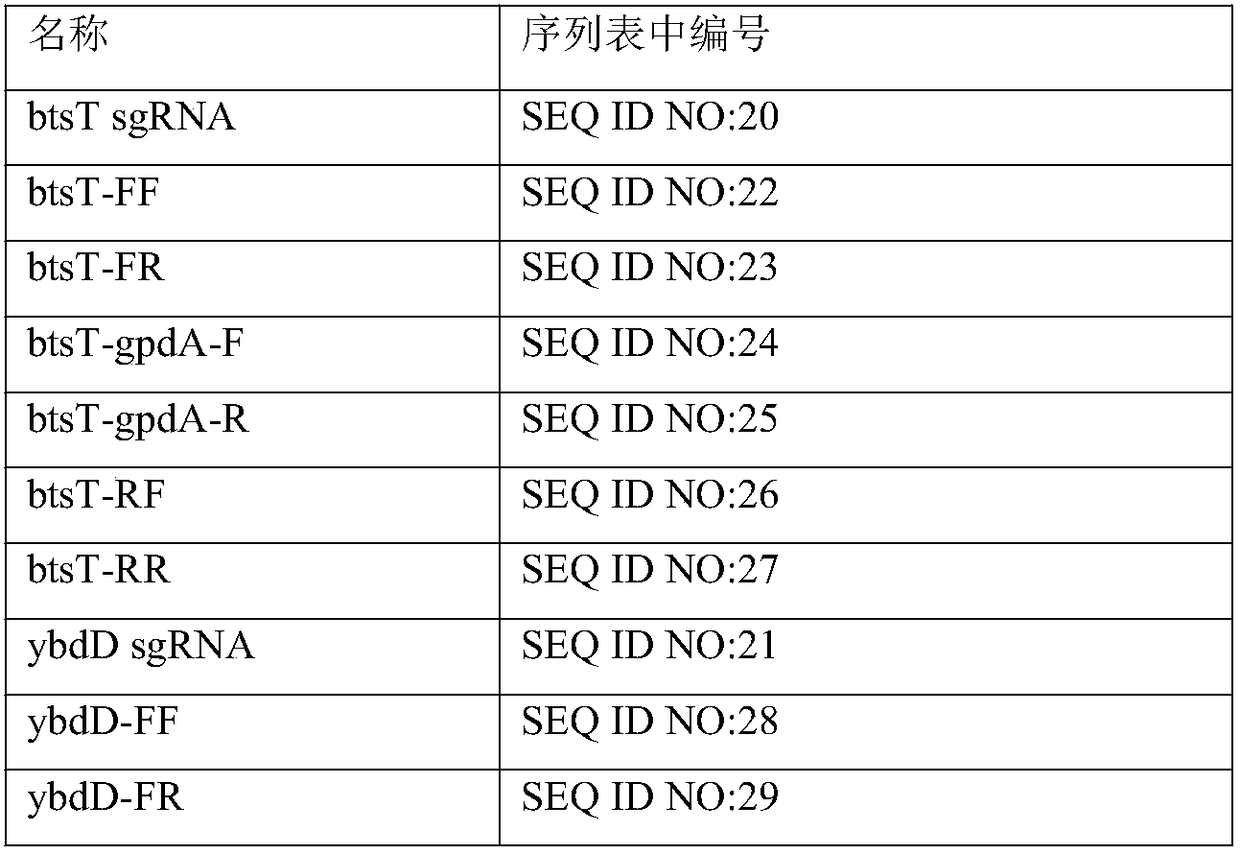
[0067] hpaD and mhpB on Escherichia coli BL21 (DE3) were single or double knockout. Among them, the gene knockout plasmid used in the present invention is pCasRed and pCRISPR-gDNA (hpaD sgRNA) and the homology arm (hpaDdonor) are introduced into Escherichia coli BL21 (DE3), and Cas9 / sgRNA induces the host to generate double strands at the hpaD gene locus Break, the recombinase Red integrates the hpaD donor into the hpaD gene to achieve gene knockout, and sequence verification. hpaD sgRNA, hpaD donor, mhpB sgRNA, and mhpB donor are respectively shown in the sequence listing SEQ ID NO: 9, SEQ ID NO: 10, SEQ ID NO: 11, and SEQ ID NO: 12. mhpB was knocked out in the same way.
[0068] Configure a solution with a pH of 7, 4 g / L of levodopa or D-danshensu, 200 g / L of wet bacteria, and measure the concentration after standing at 35 ° C for 10 hours. Table 1 shows the reaction system, levodopa and D-danshensu - The remaining amount of Danshensu.
[0069] Table 1 Residual concentra...
Embodiment 2
[0073] For the screening of L-α-amino acid transaminase, multiple L-α-amino acid transaminase genes were cloned from Escherichia coli and Lactobacillus, and expressed in Escherichia coli BL21 (DE3). When acid is the acceptor, compare the activity of various enzymes, and measure L-α according to the method described in the literature (Asymmetric synthesis of aromatic L-amino acids catalyzed by transaminases. Bioengineering Journal, 2012, 28 (11): 1346-1358.) - the activity of amino acid transaminase, the results are shown in Table 2. Therefore, it is the best to choose L-α-amino acid transaminase ect1 derived from Escherichia coli for transamination and deamination of levodopa.
[0074] Induced expression method: the recombinant Escherichia coli is transferred to LB fermentation medium (peptone 10g / L, yeast powder 5g / L, NaCl 10g / L) in the amount of 2% by volume ratio, when the cell OD 600 After reaching 0.6-0.8, add IPTG with a final concentration of 0.4mM, induce expression a...
Embodiment 3
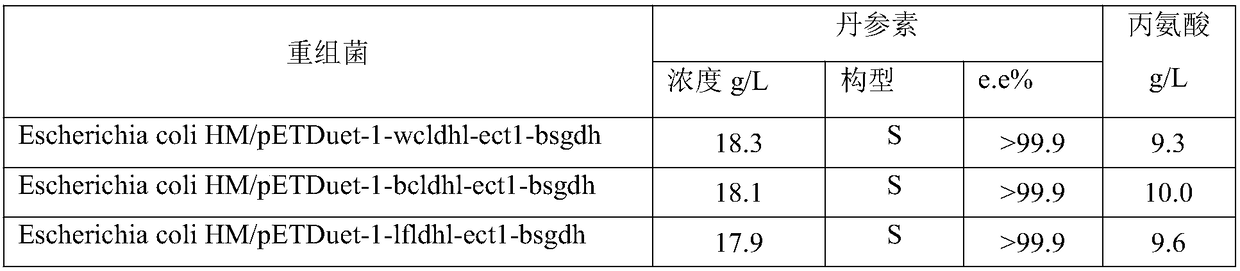
[0078] Recombinant Escherichia coli construction: first, the genes encoding L-α-amino acid transaminase, α-hydroxycarboxylic acid dehydrogenase and glucose dehydrogenase were connected to the plasmid. A recombinant plasmid co-expressing the three genes was obtained, and the plasmid was transformed into Escherichia coli HM, and positive transformants were obtained by screening on an antibiotic plate, that is, recombinant Escherichia coli was obtained.
[0079] After the expression of recombinant Escherichia coli is induced and expressed, the bacterial cells are collected, and in a reaction volume of 100ml, the cell wet weight is 40g / L, levodopa is 40g / L, pyruvate is 30g / L, glucose is 30g / L, pH 8.0, and reacted at 35°C , time 12 hours. After the conversion, the yield and configuration of Danshensu were determined by liquid chromatography.
[0080] Table 3 Comparison of various recombinant bacteria
[0081]
[0082]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com