A probe combination and method suitable for physical location of rDNA in plant chromosomes
A technology for physical positioning and chromosomes, which is applied in the field of probe combinations for physical positioning of plant chromosome rDNA, can solve the problems of high reagent cost, cumbersome process, inconsistent length, etc., and achieves the effects of convenient physical positioning, simple and convenient operation steps, and improved sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] This embodiment provides a probe combination for plant chromosome in situ hybridization, which includes a 5S rDNA probe and a 45S rDNA probe;
[0048] The nucleotide sequence of the 5S rDNA probe is shown in SEQ ID NO.1;
[0049] 45S rDNA probe consists of 5.8S rDNA probe, 18S rDNA probe and 25S rDNA probe.
[0050] The nucleotide sequence of the 5.8S rDNA probe is shown in SEQ ID NO.2, the nucleotide sequence of the 18S rDNA probe is shown in SEQ ID NO.3, and the nucleotide sequence of the 25S rDNA probe is shown in SEQ ID NO .4 shown.
[0051] Among them, both ends of the 5S rDNA probe are labeled with FAM fluorescent groups;
[0052] The 5.8S rDNA probe, 18S rDNA probe and 25S rDNA probe are all labeled with TAMRA fluorophore at both ends.
[0053] The nucleotide sequences and modifications of each rDNA oligonucleotide probe are shown in Table 1 below:
[0054] Table 1 Nucleotide sequence and modification of each rDNA probe
[0055]
[0056]
Embodiment 2
[0058] A kind of method for plant chromosome in situ hybridization that the present embodiment provides, it comprises the steps:
[0059] 1 Preparation of metaphase chromosomes
[0060] (1) Material collection: take the root tips (0.5-1.5 cm) of vigorously growing plants as materials.
[0061] (2) Pretreatment: pretreat the freshly obtained root tip with 0.7mM cycloacetamide at room temperature for 4-8 hours (the time required for different species is different, determined according to the actual situation), rinse with distilled water, and transfer to the card Nuo fixative (3:1 ethanol:acetic acid, v / v).
[0062] (3) Enzymolysis: place the fixed root tip in distilled water and rinse thoroughly, then cut the root tip meristem, and put it into an enzyme mixture containing 4% cellulose and 2% pectinase. The enzymatic hydrolysis is about 0.5-1.5h in a 37°C incubator (the time required for different species is different, and it is determined according to the actual situation).
...
Embodiment 3
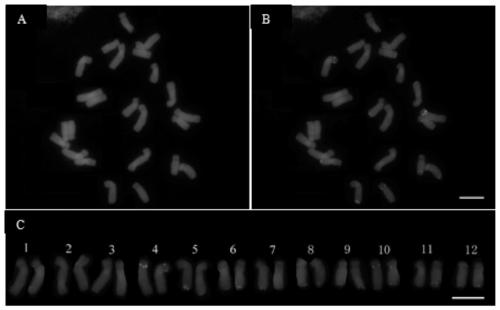
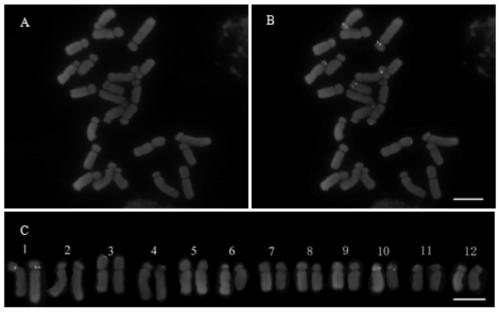
[0082] In this example, Ili tulip (T.iliensis Regel) of the genus Tulip was used as a material, and fluorescence in situ hybridization was carried out according to the method in Example 2. The results were as follows: figure 1 shown.
[0083] From figure 1 It can be seen that the chromosomes are in good shape and the fluorescence signal is very clear. There are 5 5SrDNA loci (green) in Yili tulip, 4 of which are located in the near centromere region on the short arms of homologous chromosomes 4 and 10, and the other 1 One is located on the long arm of one of the homologous chromosomes 11. There are 15 45S rDNA loci (red) in Yili tulip, most of which are located at the end of the long arm, 2 of which are located at the end of the short arm of chromosome 11, and 12 loci are located at 5, 6, 8, 9, 10 and 12 end of the long arm of the homologous chromosome. One of the homologous chromosomes 7 has a 45S rDNA locus at the end of the long arm, and the other has no signal.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com