Rapid detection method of live virus and application of rapid detection method
A detection method and technology for live virus, applied in the field of rapid detection of live virus, to achieve the effect of verifying the effectiveness of virus inactivation process, good quantitative measurement effect, and rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
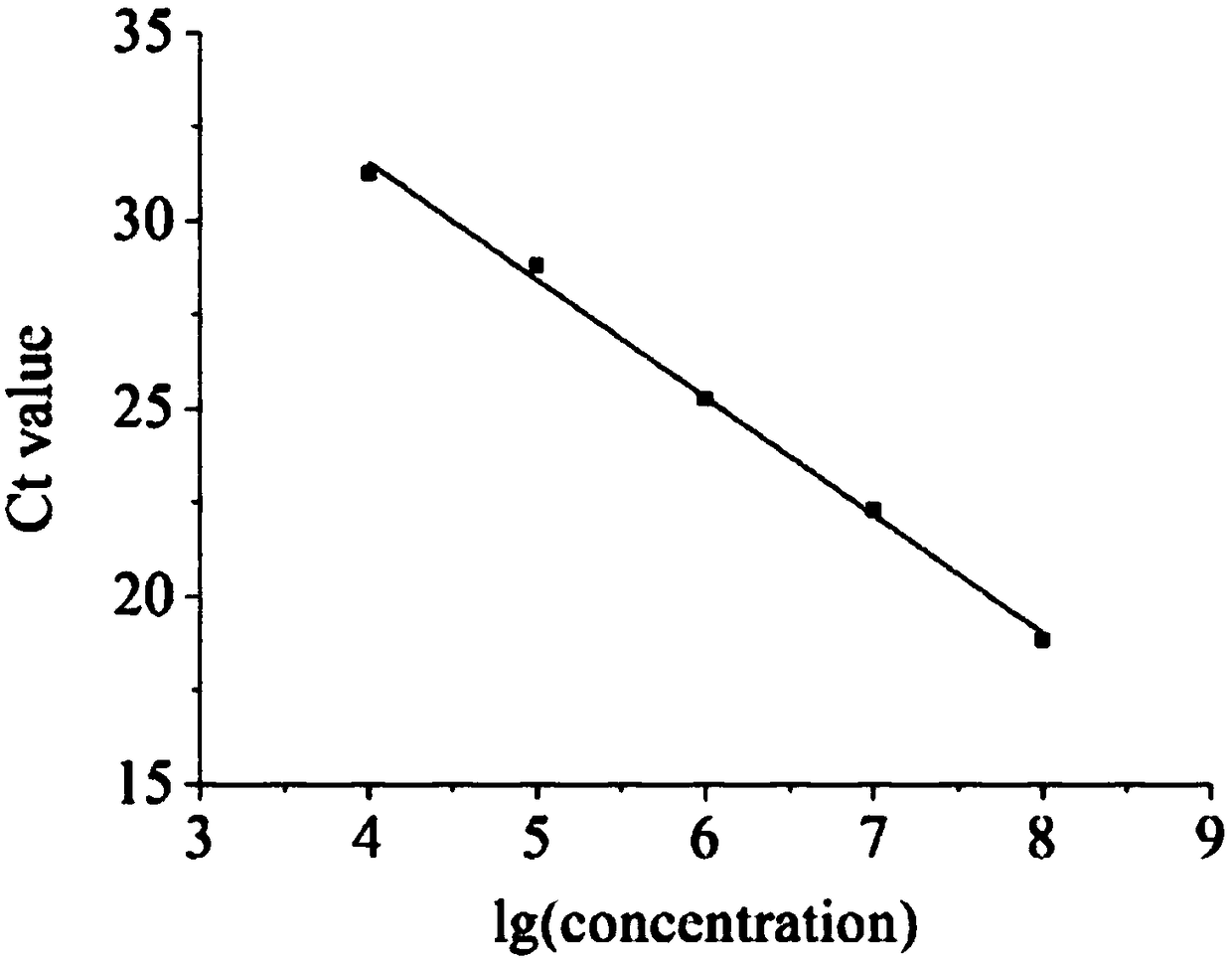
[0061] Establishment and verification of a rapid detection method for live virus (ie, ICC-qPCR method).
[0062] 1. Establishment and verification of qPCR method.
[0063] 1. Preparation of plasmid DNA standards.
[0064] The strain category of PRV (pseudorabies virus) is Barthar strain, and gB is a gene name of this PRV strain. Referring to the gB gene sequence of Barthar strain in GeneBank, after experimental screening and verification, the following primer set was selected as the primers for this experimental research, and the primer sequences are:
[0065] FP: 5'-GTCACCTTGTGGTTGTTG-3' (SEQ ID No. 1)
[0066] RP: 5'-CCACATCTACTACAAGAAC G-3' (SEQ ID No.2)
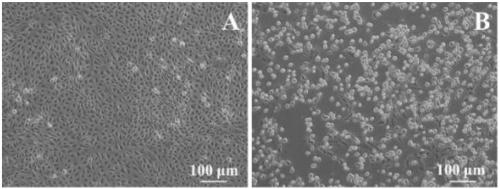
[0067] The fragment amplified with the above primer set was 180 bp in length. The electrophoresis product of the amplified fragment was recovered and purified, and then connected to the pMDTM18-T cloning vector for cloning. The positive clones were initially identified by nucleic acid electrophoresis and qPCR. The ide...
Embodiment 2
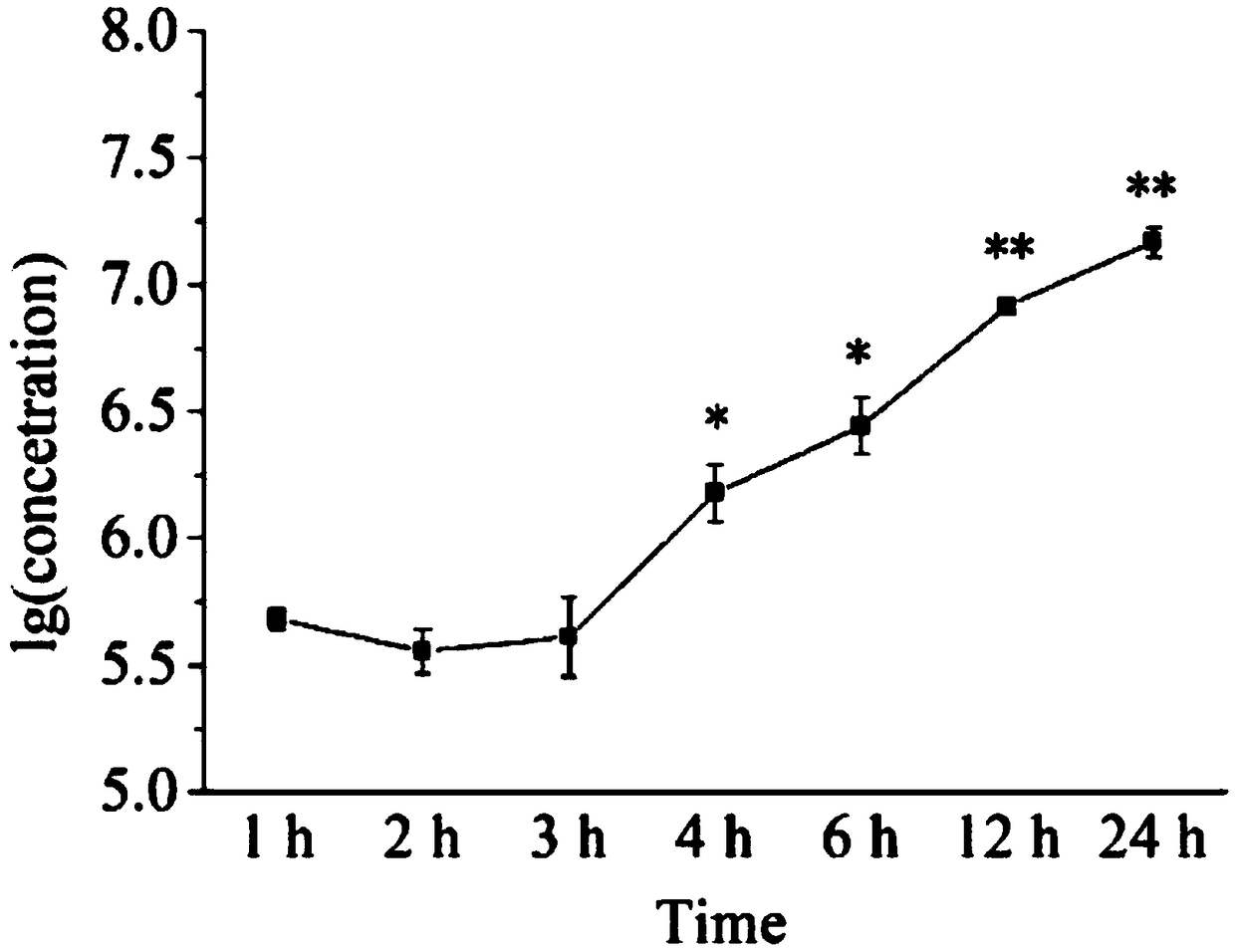
[0113] The application of the ICC-qPCR detection method of PRV in the validity verification of a virus inactivation process (electrophysiological catheter ethylene oxide virus inactivation process).
[0114] 1. Pollutant load.
[0115] Based on the physical characteristics of the electrophysiological catheter, clinical application, and the characteristics of the pollutants, the pollutants in the whole blood test were selected (100 mL of citrated sheep whole blood, 50 mL of calf serum, and 50 mL of normal saline, and 2 mol / L CaCl was added before use). 2 0.01mL), to simulate the contamination after clinical use. A high titer of PRV virus was added to the whole blood test contaminant to prepare the whole blood test contaminant containing PRV virus.
[0116] Using 12 sterile solid electrophysiological catheters, by measuring the length (L) and diameter (2r) of the shaft of the catheter for experimental contamination (measured with a vernier caliper), calculate the approximate s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com