CPA primer, kit and detection method for escherichia coli O157:H7
A technology of Escherichia coli and O157, which is applied in biochemical equipment and methods, microbial measurement/inspection, recombinant DNA technology, etc., can solve the problems of low sensitivity and low detection sensitivity, achieve low detection cost, simple and fast operation, and ensure reliability effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] The method for detecting escherichia coli O157:H7 based on cross-primer constant temperature amplification reaction technology may further comprise the steps:
[0053] 1. The method for detecting pathogenic microorganisms based on cross-primer constant temperature amplification reaction technology, the present embodiment takes Escherichia coli O157:H7 as an example, and the reagents used are as follows:
[0054] a. The stripping primers 4s and 5a, the cross-amplification primers 2a1s, and the specific primers 2a and 3a each at a concentration of 10 μM, the primer sequences are as shown in the preceding SEQ ID NO.1-SEQ ID NO.5;
[0055] b.2×Reaction stock solution: Tris-HCl with concentration of 40.0mM, ammonium sulfate of 20.0mM, potassium chloride of 20.0mM, magnesium sulfate of 16.0mM, Tween 20 of 0.2% (v / v), 1.4M Betaine, 10.0mM dNTPs (each) composition;
[0056] c. Bst DNA polymerase (large fragment, NEB company) aqueous solution with a concentration of 8U / μL;
[...
Embodiment 2
[0068] The cross constant temperature amplification reaction detects the optimum amplification time test of Escherichia coli O157:H7, comprising the following steps:
[0069] According to the reaction system of Example 1, the cross constant temperature amplification method was constructed, and different amplification time tests of 10 minutes, 20 minutes, 30 minutes, 40 minutes, 50 minutes, 60 minutes, 70 minutes, 80 minutes, and 90 minutes were carried out to determine Optimal amplification time.
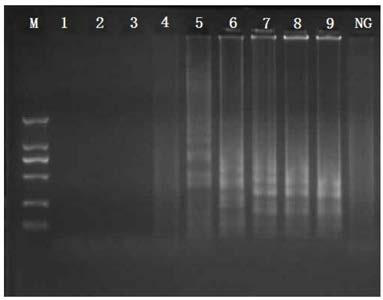
[0070] The reaction system was constructed according to Example 1, sterile water was used as a blank control, and 2% agarose gel electrophoresis was performed on the amplified product.
[0071] The result is as figure 2 As shown, when the reaction time is 40 minutes to 90 minutes, a ladder-like band appears, and there is no significant difference in the ladder-like band from 60 minutes to 90 minutes, so the cross-primer constant temperature amplification reaction time can be reduc...
Embodiment 3
[0073] The cross constant temperature amplification reaction detection Escherichia coli O157:H7 specificity test comprises the following steps:
[0074] The genomic DNA of Escherichia coli O157:H7ATCC43895 and non-Escherichia coli was established according to the reaction system and conditions in Example 1. A cross-isothermal amplification reaction detection method was carried out to carry out a specificity test;
[0075] Among them, non-Escherichia coli are: Salmonella ATCC29629; Salmonella ATCC19585; Salmonella ATCC14028; Salmonella ATCC13076; Listeria monocytogenes ATCC19116; Listeria monocytogenes ATCC19114; Species ATCC19113; Pseudomonas aeruginosa ATCC27853; Staphylococcus aureus ATCC27664; MRSA NCTC10442; MRSA N315; MRSA 85 / 2082; Methicillin Staphylococcus aureus CA05; Vibrio parahaemolyticus ATCC17802; Vibrio parahaemolyticus ATCC27969; Lactobacillus casei BM-LC14617.
[0076]Set the Escherichia coli O157:H7 genome as the positive control, and the nucleic acid-free wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com