Method for screening and characterizing metabolite-protein interaction system
A metabolite and protein technology, applied in the field of analytical chemistry and biochemistry, which can solve the problem of not being able to screen metabolites in large quantities
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
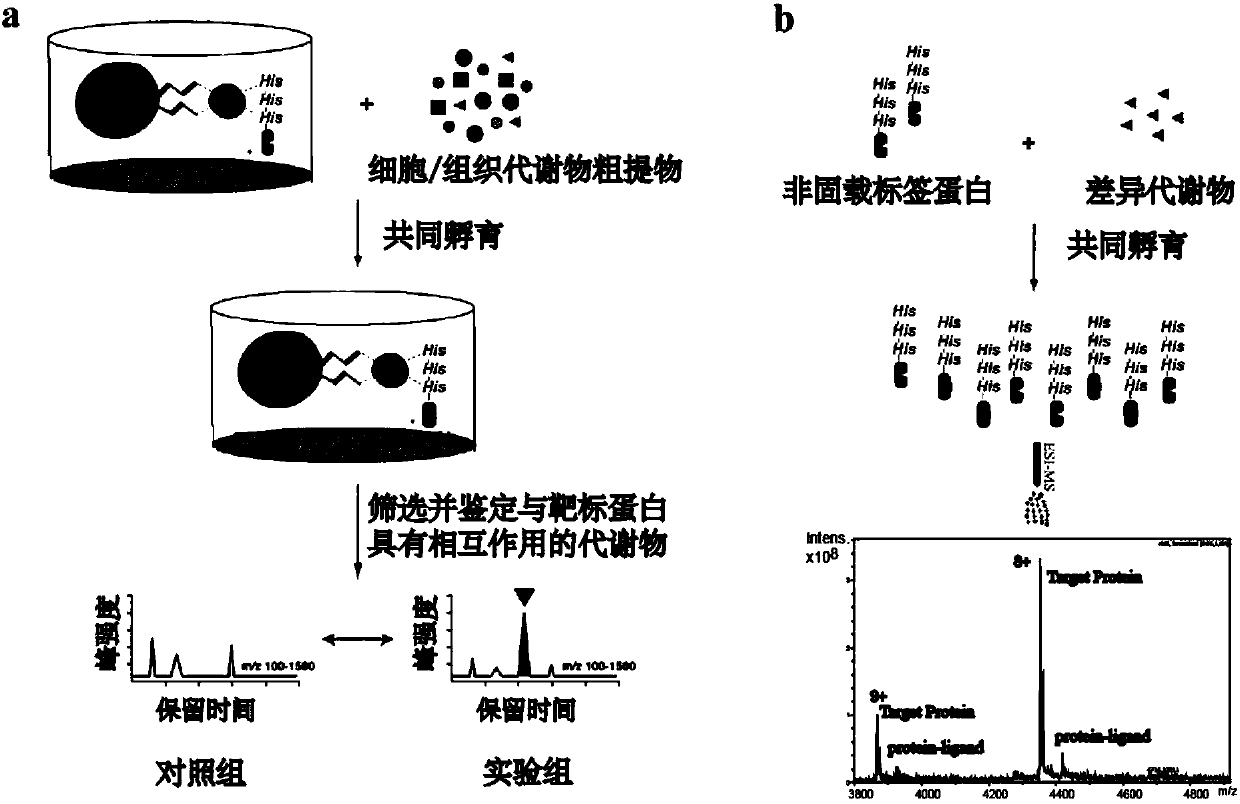
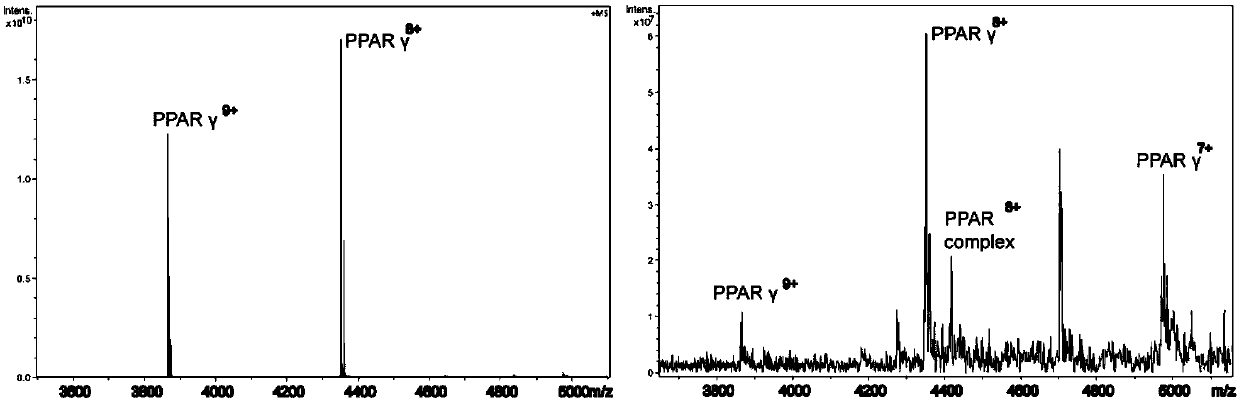
[0022] Interaction between peroxidase value-proliferating receptor γ and endogenous metabolites based on metabolomics and structural mass spectrometry
[0023] (1) Heterologous overexpression of catalase proliferating receptor protein gamma (PPAR-gamma) with histidine (His6-) tag in Escherichia coli. Using the histidine tag on the PPAR-γ, the Escherichia coli was crushed and then affinity-purified using a nickel column: washing with 20mM imidazole buffer for 3-4 times, and elution with 500mM imidazole buffer.
[0024] (2) Use lipid extraction solution (2ml chloroform, 2ml methanol, 1ml water) to extract small lipid molecules from 10mg brain tissue, take the supernatant and freeze-dry it with 4% (DMSO / EtOH=1:1) Reconstituted in PBS buffer as brain tissue lipid extract. Immobilize 140 μL of the PPAR-γ protein purified in the previous step on a 20 μL nickel resin column in the form of affinity immobilization, centrifuge to remove the supernatant, and add brain tissue lipid extra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com