Screening method for real-time fluorescent quantitative PCR internal reference gene of Hibiscus hibiscus
A technology of real-time fluorescence quantification and internal reference genes, applied in the fields of genomics, instrumentation, sequence analysis, etc., can solve the problems that there are no related reports on the internal reference genes of Hibiscus hibiscus
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1. Selection of internal reference genes
[0081] The present invention selects 10 candidate internal reference genes from the existing Hibiscus transcriptome database, and designs real-time fluorescent quantitative PCR specific primers for each internal reference gene.
[0082] Comparison and analysis of candidate internal reference gene sequences: Submit the candidate gene sequences to BLASTX in NCBI for comparison and analysis. The sequence identity of each internal reference gene sequence in Hibiscus hibiscus with homologous genes of related species is more than 85%, and most All above 90%. The sequence identity with the model plant Arabidopsis is more than 83%. The comparison results are shown in Table 1.
[0083] sheet BLASTX comparative analysis of internal reference genes
[0084]
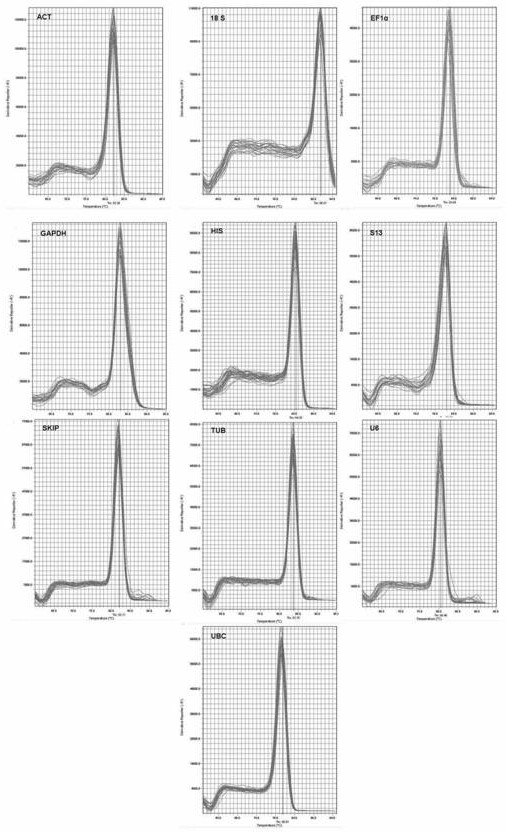
[0085] Design and verification of real-time fluorescent quantitative PCR primers: using the candidate gene sequence as a template, use GenScript’s online primer desig...
Embodiment 2
[0089] Embodiment 2. Real-time fluorescence quantitative PCR experiment
[0090] The experimental materials were divided into two groups. One group was the tender leaves of sea hibiscus, which were treated with 7 kinds of abiotic stresses including drought, high salt, low temperature, high temperature, abscisic acid, salicylic acid and methyl jasmonate 0, 1, 6, 12, 24 and 48 h; the other group is the tissues and organs, which are the young roots, old leaves, young leaves, receptacles, petals, and stamens of 10-year-old sea hibiscus. The collection of plant treatment materials is as follows: one group of materials used seashore hibiscus seedlings as materials, and simulated drought and high-salt stress with 15% polyethylene glycol 6000 and 400 mM sodium chloride respectively; the seashore hibiscus seedlings were placed in 4°C C and temperature 42°C incubator to simulate low temperature and high temperature stress. The conditions of hormone treatment were as follows: spray Hibi...
Embodiment 3
[0094] Example 3. geNorm, NormFinder and BestKeeper software evaluation of internal reference genes under all samples
[0095] geNorm software
[0096] In the present invention, the download address of the geNorm software is https: / / genorm.cmgg.be / , and its steps and standards are as follows:
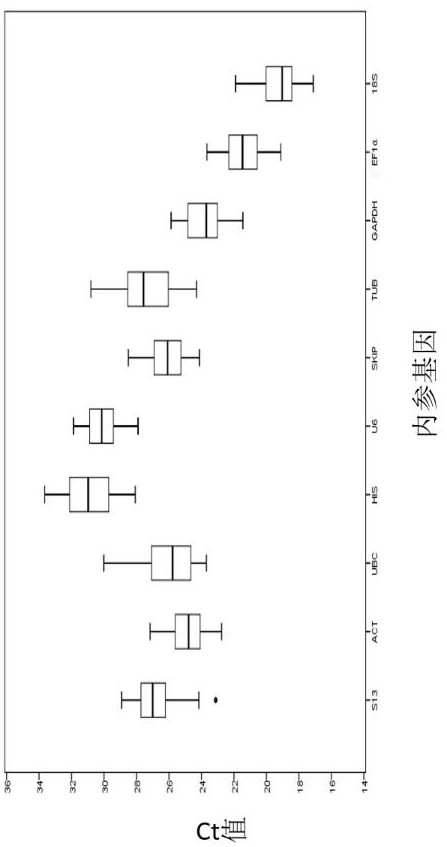
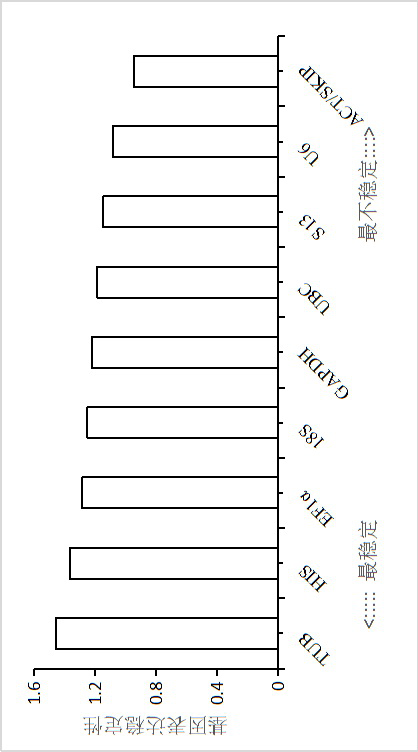
[0097] In the present invention, geNorm first realized the ranking of the expression stability of 10 internal reference genes by calculating the average expression stability (M), and the results showed that in all samples, the two most stable internal reference genes were actin ( ACT ) and SKI-associated proteins ( SKIP ) ( image 3 ), the most unstable is tubulin ( TUB ), the stability ranking of each internal reference gene is as follows: Actin ( ACT ) and SKI-associated proteins ( SKIP )> small nuclear RNA U6 ( U6 )> ribosomal protein S13 ( S13 ) > ubiquitin-conjugating enzyme E2 ( UBC )>Glyceraldehyde-3-phosphate dehydrogenase ( GAPDH )>18S ribosomal RNA ( 18S ) > ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com