QLAMP (quantitative loop mediated isothermal amplification) method for quantitatively monitoring infection and propagation of Chinese walnut dry rot and used primers
A pecan dry rot and quantitative loop technology, applied in the fields of plant disease epidemiology, dynamic monitoring and early warning, biotechnology and plant disease epidemiology, can solve the problems of complicated detection process, low detection sensitivity, and long detection time, etc. Achieve the effect of stable reaction, high sensitivity and optimized reaction temperature
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
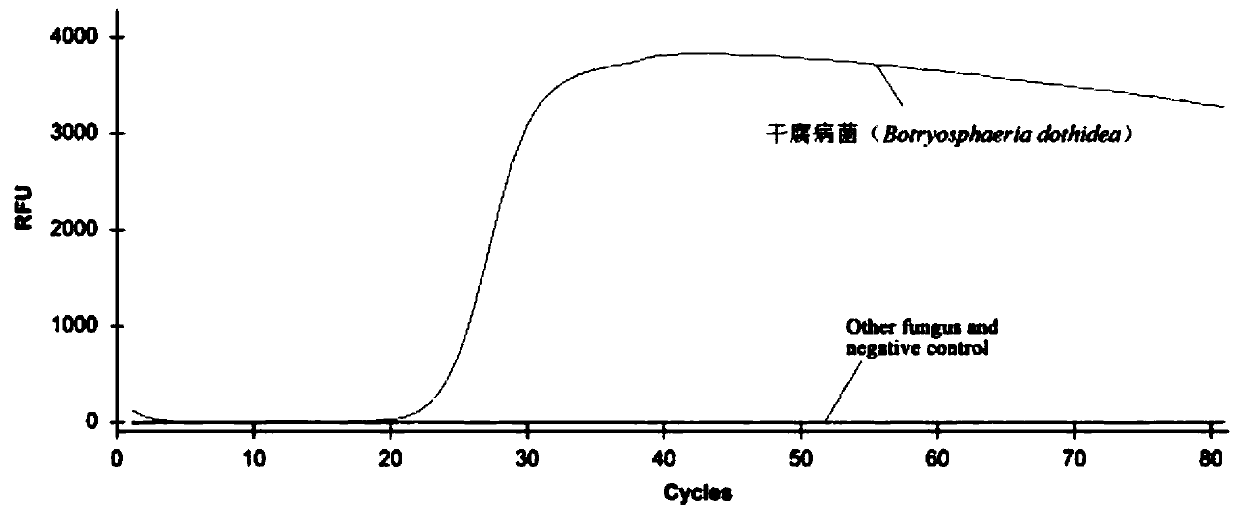
[0077] Embodiment 1, the specificity analysis of detection system
[0078] Botryosphaeria dothidea was used as a positive control, Trichoderma viride, Fusarium proliferatum, Pestalotiopsis microspora, Nigrospora chinensis, chain Alternaria regonensis and Daldinia sp. were used as negative controls, and double distilled water was used as blank control for testing.
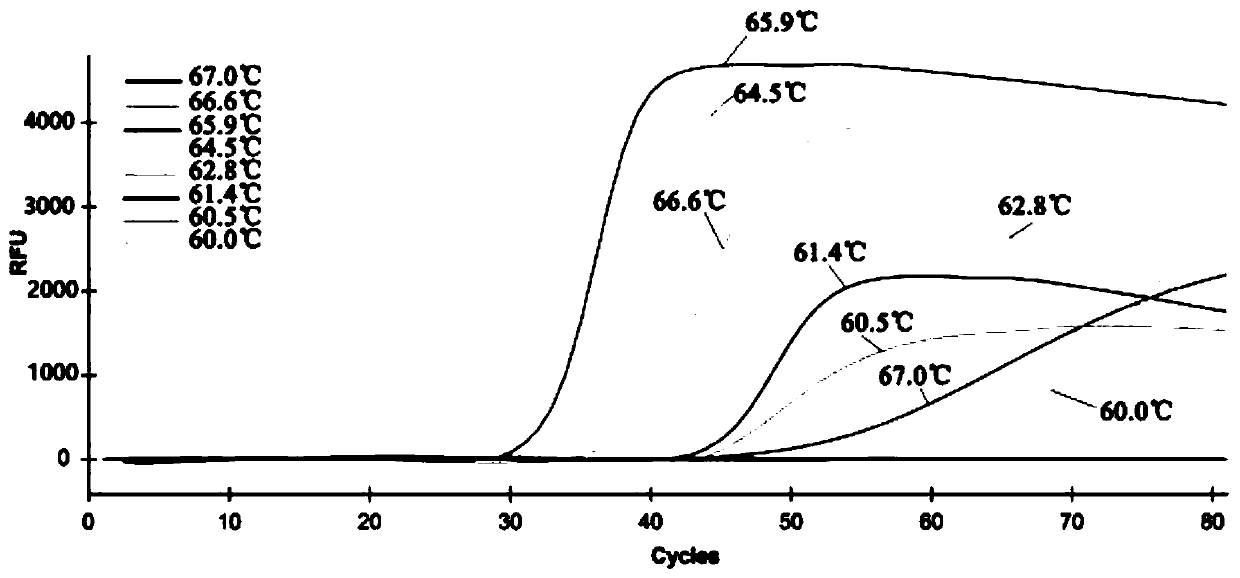
[0079] A total of 19 μL of the detection solution was added, and 1 μL of the DNA template to be tested (concentration between 0.001 ng / μL and 100 ng / μL, both positive and negative controls, 0.1 ng / μL) was added to form a 20 μL detection reaction system. The 20 μL system contains 8 U / μL BstDNA polymerase 1 μL, 10×ThermoPol Buffer 2.0 μL, 20 μM FIP primer 1.6 μL, 20 μM BIP primer 1.6 μL, 10 μM F3 primer 0.25 μL, 10 μM B3 primer 0.25 μL, 25 mM MgCl 25.0 μL, 2.0 μL of 10 mM dNTPs, 2.4 μL of 5M betaine, 1.2 μL of 10×SYBR Green I, 1 μL of DNA template, and make up to 20 μL with double distilled water. The q-LAMP amplif...
Embodiment 2
[0081] Embodiment 2, the sensitivity detection of primer
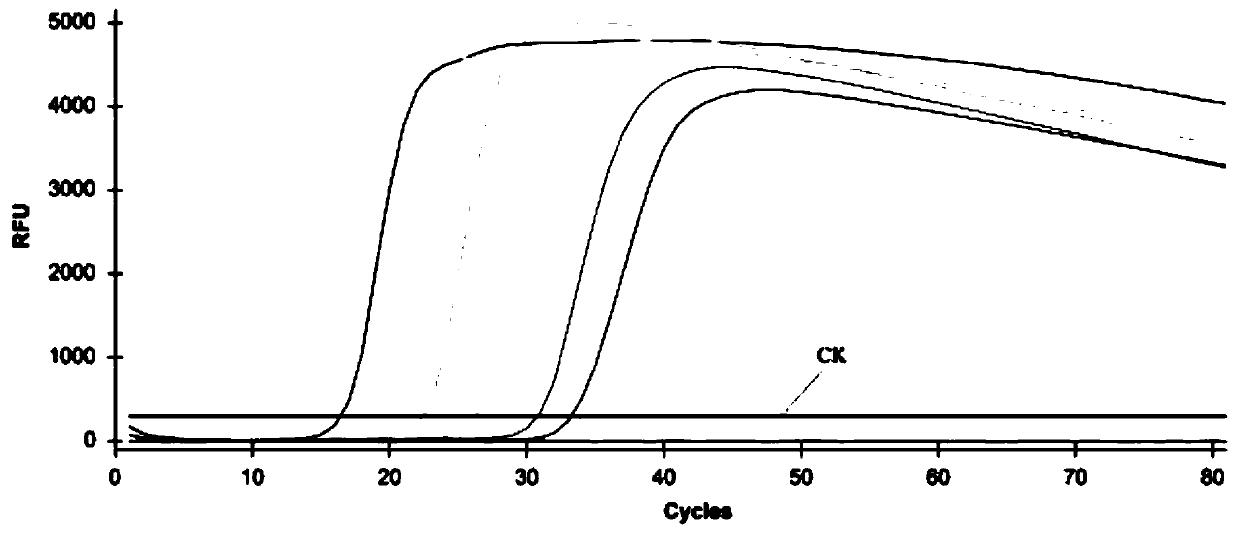
[0082] With the dry rot pathogen q-LAMP detection system after optimization (as described in embodiment 1) to the DNA solution detection result of the dry rot pathogen pure hyphae of gradient dilution as follows Figure 4 As shown, when the DNA template concentration is 100ng / μL, 10ng / μL, 1ng / μL, 0.1ng / μL, 0.01ng / μL and 0.001ng / μL, it can be effectively amplified, and the repeatability of the same sample is reliable. The amplification time corresponding to each concentration is 16.32min, 18.68min, 22.43min, 26.63min, 30.84min and 33.26min respectively. The results showed that the theoretical detection line of dry rot pathogen DNA was 1pg DNA in the established q-LAMP detection system.
Embodiment 3
[0083] Embodiment 3, the acquisition of standard curve:
[0084] Standard curve 1, when the spore concentration is 1.0×10 5 pcs / mL, 1.0×10 4 pcs / mL, 1.0×10 3 pcs / mL, 1.0×10 2 / mL and 1.0×10 1 When samples / mL, the amplification time corresponding to each concentration is 16.67min, 21.51min, 28.69min, 36.26min and 43.61min, according to the dilution sample C T Standard curve between the value (y) and the Log value (x) of the number of spores per milliliter of sample: y=-6.863x+49.937, correlation coefficient R 2 =0.9922, the calculation formula for the number of spores (pieces) is
[0085] Calculation formula: construct a standard curve, use the Log value of the number of spores per milliliter as the x-axis, and the corresponding C T The value is plotted on the y axis, showing a decreasing linear equation [y=mx+b, or y=m (log number of spores per milliliter of sample)+b], the following equation can be deduced from the decreasing linear equation to determine Unknown samp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com