Anti-H7N9 whole human monoclonal antibody 5G22, and preparation method and application thereof
A monoclonal antibody, fully human technology, applied in the field of immunology, can solve the problems of rimantadine drug resistance and no effective treatment methods, and achieve the goal of reducing cumbersome operations and costs, high affinity and specificity, and low production costs Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] (1) Construction of NTH-3T3 cell line stably expressing CD40L
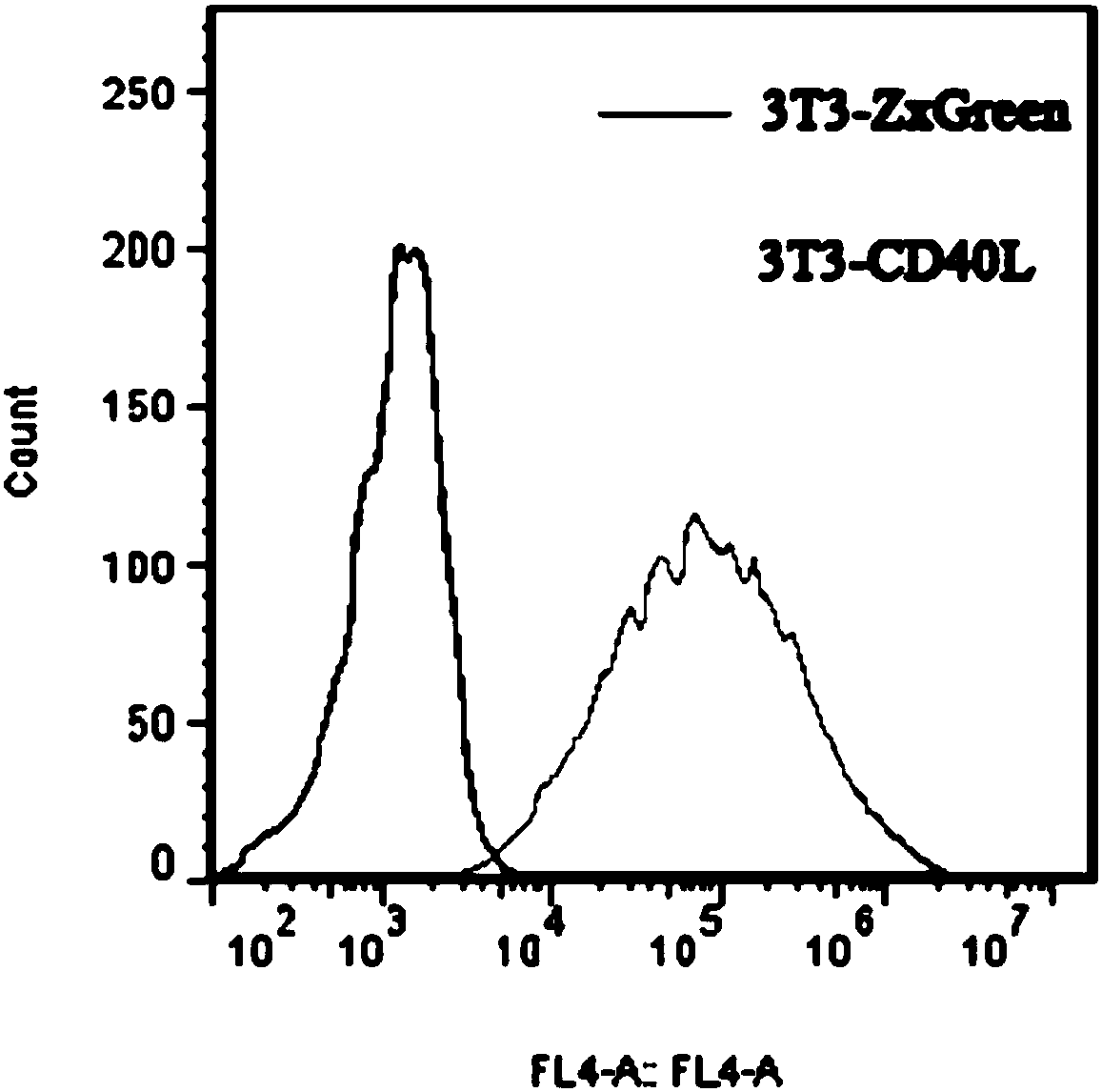
[0044] 3T3-CD40L feeder cells were established using lentivirus. The lentiviral expression vector pLVX-CD40L was constructed, transfected into 293T cells, and the virus supernatant was collected on the fourth day of transfection. NIH-3T3 cells were activated, cultured for 3 generations, infected with lentivirus, continued to be cultured and passed 3 times. Use a flow cytometer to sort the cells whose FITC fluorescence intensity is near the MFI, and add them back to the culture flask at 37°C, 5% CO 2 Cultivate and detect in the incubator, and the test results are as follows: figure 1 As shown, 3T3 cells expressing CD40L and 3T3 cells transfected with empty vector pLVX (with ZxGreen) were stained with anti-CD40L with APC, and then analyzed by flow cytometry. It was found that all 3T3-CD40L feeder cells expressed CD40L. When the cells grow to 80%-90%, digest and collect the cells at a concentration of 1×10...
Embodiment 2
[0066] Example 2 Cloning, recombination and expression of humanized monoclonal antibody 5G22 gene
[0067] The B cells obtained in Example 1 capable of secreting the antibody 5G22 binding to the H7N9 virus were lysed, and the lysate was taken for reverse transcription of RNA to obtain the PCR template cDNA of the human antibody gene. The heavy chain and light chain genes of the antibody were cloned using cDNA as a template, and recombinantly expressed and purified in eukaryotic 293F or HEK293 cells.
[0068] specifically:
[0069] (1) Transfer the B cell liquid to a 96-well plate (Eppendorf, 030133366).
[0070] (2) Reverse transcription system: 150ng random primer (invitrogen, 48190-011), 0.5ul 10mM dNTP (Invitrogen, 18427-088), 1μl 0.1M DTT (Invitrogen, 18080-044), 0.5% v / v Igepal CA -630 (Sigma, I3021-50ML), 4U RNAsin (Promega), 6U Prime RNAse Inhibitor
[0071] (Eppendorf) and 50U III reverse transcriptase (Invitrogen, 18080-044), add DEPC water to 14μl / well.
[0072...
Embodiment 3
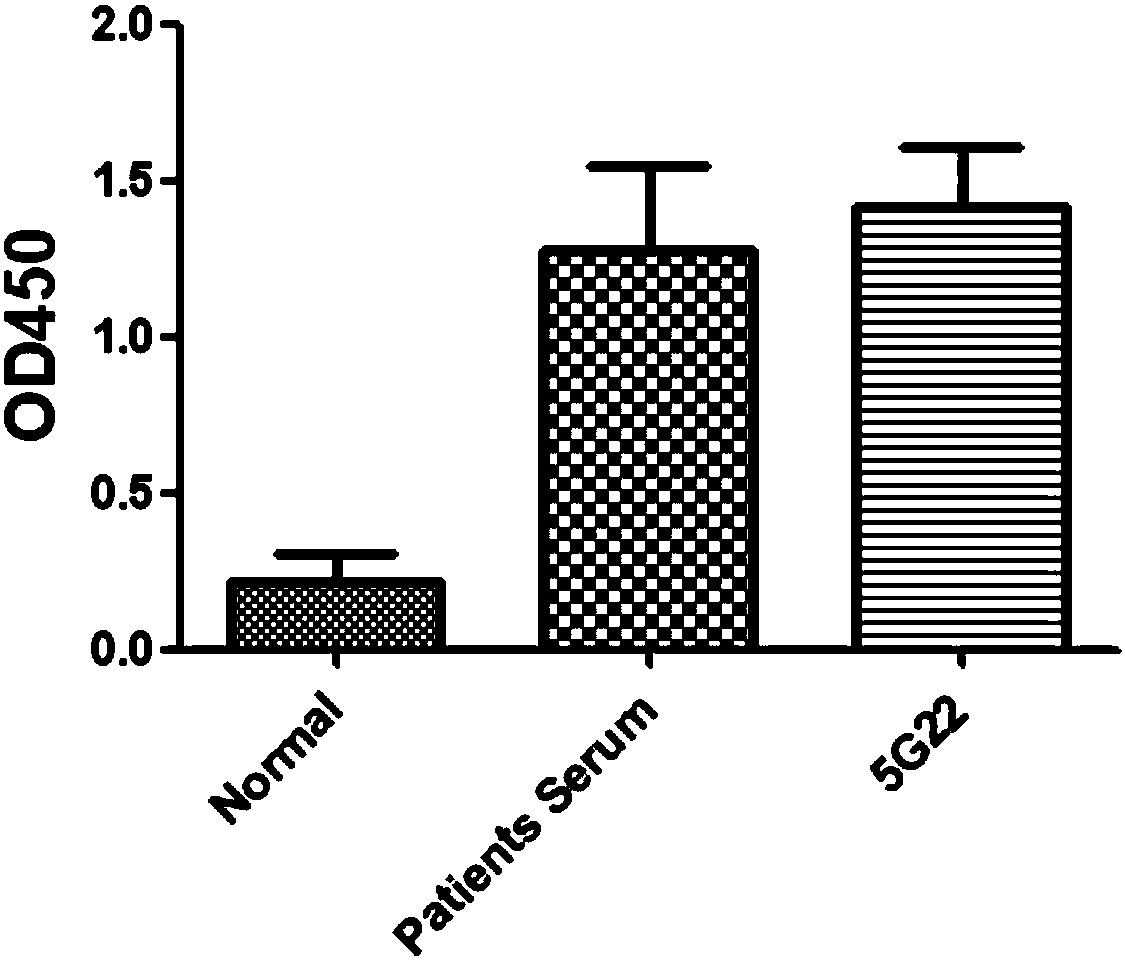
[0093] Example 3 In vitro neutralization experiment of humanized monoclonal antibody 5G22
[0094] Using the virus-infected cell model (canine kidney cell MDCK), the inhibitory effect and effect of the antibody on the H7N9 influenza virus with PR-8 as the backbone were evaluated by microneutralization-ELISA experiment, and the anti-influenza virus activity of the antibody was detected. The specific operation is as follows:
[0095] 1 cell plating
[0096] Digest logarithmic growth phase cells with trypsin, collect by centrifugation after termination, blow evenly, and prepare single cell suspension;
[0097] Adjust the cell concentration to 5×104 cells / ml with cell culture medium, inoculate in 96-well cell culture plate, and store the cells at 37°C and 5% CO 2 Incubate overnight in the incubator.
[0098] 2 Antibody and virus pretreatment
[0099] 5G22 antibody set up 10 concentration gradients, followed by 10-10 10 Three-fold dilutions were performed, and three parallel w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com