Sample pretreatment process suitable for nucleic acid detection test strip for aquatic diseases
A technology for sample pretreatment and detection of test strips, applied in the biological field, can solve the problems that samples cannot be used, nucleic acid test strips are false negative, and nucleic acid detection test strips cannot be popularized and applied, so as to improve the detection effect, simplify nucleic acid extraction and Effects of purification steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] (1) Streak inoculate Vibrio splendidus on 2216E solid medium plate and culture at 28°C for 24 hours, pick a single colony in 2216E liquid medium, culture overnight at 28°C with 200rpm shaking, and dilute the culture solution with normal saline to Concentrations were 1×10 6 , 1×10 5 , 1×10 4 , 1×10 3 , 1×10 2 , 1×10 CFU / mL, take 1 mL of bacterial culture solution, centrifuge at 8000 r / min for 5 min, discard the supernatant, add lysate (50 mM Tris-HCl, 10 mM EDTA, 10 mM NaCl, 1% SDS, 0.1 mg / L proteinase K) 1 mL, in a 70°C water bath for 10 min.
[0044] (2) Centrifuge the above treatment solution at 12,000 rpm for 5 minutes, take 800 μL of the supernatant and add it to a type A nucleic acid adsorption column, and place it for 2 minutes. Centrifuge at 12000 rpm for 1 min, discard the liquid in the collection sleeve, and repeat this step once;
[0045] (3) Add 1 mL of 80% ethanol (pH 5.5), and centrifuge at 12000 rpm for 1 min;
[0046] (4) Leave it open for 3 minute...
Embodiment 2
[0050] (1) Cut 0.5 g of the body wall tissue of sea cucumber into pieces, transfer it to a glass homogenizer, add 5 mL of homogenate solution (100 mM Tris-HCl, 500 mM EDTA, 20 mM NaCl, 10% SDS), and homogenize Transfer the ground homogenate to a centrifuge tube, add 20 μL of proteinase K (20mg / mL), invert and mix, place in a constant temperature water bath at 70°C for 20 minutes, and cool on ice.
[0051] (2) Centrifuge the above treatment solution at 10,000 rpm for 5 minutes, take 2.5 mL of the supernatant and add it to a B-type nucleic acid adsorption column, and place it for 5 minutes. Centrifuge at 10,000 rpm for 3 minutes, discard the liquid in the collection sleeve, and repeat this step once.
[0052] (3) Add 2.5 mL of 80% ethanol (pH 5.5), centrifuge at 12000 rpm for 1 min, and repeat this step once.
[0053] (4) Leave it open for 3 minutes, add 20 μL of deionized water to the center of the silica gel adsorption membrane, leave it for 2 minutes, centrifuge at 12000 rpm...
Embodiment 3
[0057] (1) Take 100 mL of aquaculture water samples, put them in a 100°C water bath for 10 min, and cool them down at room temperature.
[0058] (2) Centrifuge the above-mentioned treatment solution at 8500rpm for 5 minutes, take 17 mL of the supernatant and add it to a C-type nucleic acid adsorption column, and place it for 5 minutes. Centrifuge at 8500 rpm for 3 minutes, discard the liquid in the collection sleeve, and repeat this step once.
[0059] (3) Add 10 mL of 80% ethanol (pH 5.5), and centrifuge at 10,000 rpm for 1 min.
[0060] (4) Leave it open for 3 minutes, add 30 μL of deionized water to the center of the silica gel adsorption membrane, let it stand for 2 minutes, centrifuge at 12000 rpm for 1 minute, and use the obtained elution solution for subsequent detection tests.
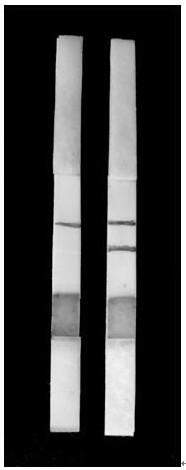
[0061] (5) Take 50 μL of the above-mentioned eluent and add dropwise on the sample pad of the Vibrio resplendent nucleic acid test strip, place the test strip in a wet box, incubate at 37°C fo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| adsorption capacity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com