Method for detecting hepatitis b virus subtype based on DNA paper-folding mark
A hepatitis B virus, virus technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of lack of visual labels in genetic information analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] Embodiment 1, the kit that detects different genotypes of virus
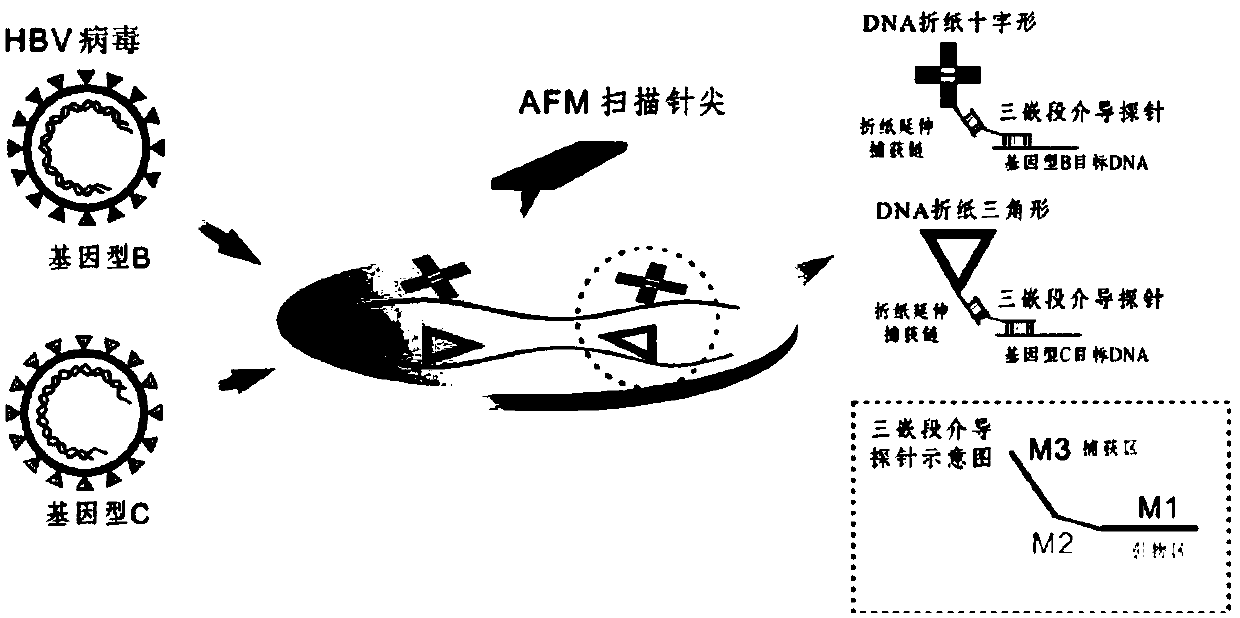
[0090] figure 1 It is a schematic diagram of directly reading DNA origami markers to identify HBV genotypes based on AFM imaging, specifically: HBV genomic DNA is extracted from serum samples, amplified by asymmetric PCR and digested with λ exonuclease, and the HBV target genome fragments are used with the corresponding DNA origami markers are used as "amplified signals" for site-specific labeling, and the corresponding markers are read directly on the AFM image to translate genotype information.
[0091] 1. Kits for detecting different genotypes of viruses
[0092] Kits for detecting different genotypes of viruses, including multiple sets of biological products;
[0093] Each set of biological products includes one or more groups of reagents;
[0094] Each set of biological products corresponds to 1 virus genotype and 1 shape of DNA origami;
[0095] Each group of reagents consists of a DNA origami w...
Embodiment 2
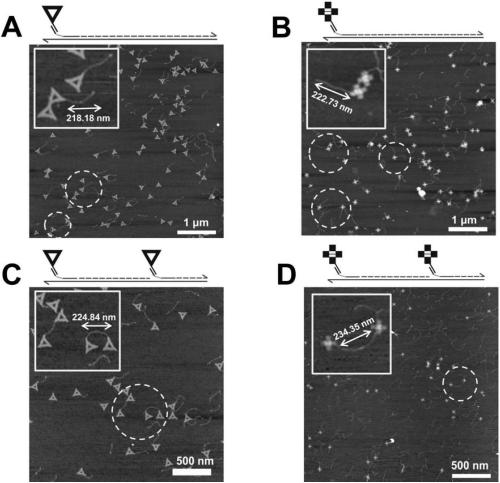
[0129] Embodiment 2, the application of triangle DNA origami and cross DNA origami in detecting HBV genotype
[0130] Figure 6 It is a schematic diagram of the structure of the DNA origami label and the position of the probe extension used in the experiment.
[0131] 1. Detection of HBV genotype by triangle DNA origami and cross DNA origami
[0132] 1. Extraction of total nucleic acid from the sample to be tested
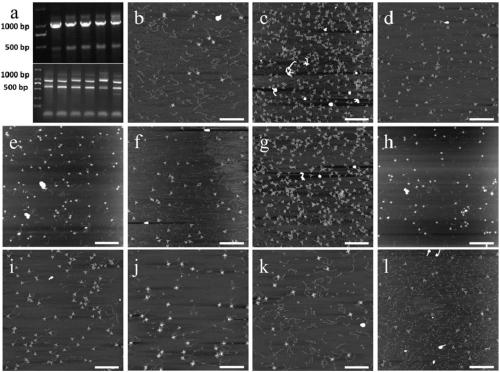
[0133] The samples in this study came from the whole blood samples of patients who were diagnosed with HBV infection by ELISA immune reaction pre-detection in Wuxi People's Hospital, of which 5 cases were pre-diagnosed as genotype B, and the other 6 cases were genotype C. Use the TIANampVirus DNA / RNAKit Viral Genome Nucleic Acid Extraction Kit to extract HBV viral DNA from whole blood samples.
[0134] The DNA was eluted in 50 μl of elution buffer (included with the kit), stored at -80° C. for future use, and the HBV viral DNA of 11 samples was obtained.
[013...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com