Fluorescent quantitative PCR method for detecting vibrio parahaemolyticus causing acute hepatic pancreatic necrosis
A technology of vibrio hemolyticus and fluorescence quantification, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism measurement/inspection, which can solve the problems of difficult detection, poor sensitivity, and long detection time of latent infection samples, etc. problem, to achieve the effect of improved stability, good quenching effect and small background
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Real-time quantitative PCR method for detection of Vibrio parahaemolyticus causing acute hepatopancreatic necrosis
[0054] 1. Design and synthesis of primers and TaqMan-MGB probes
[0055] with Vp AHPND PirA Vp The gene is the detection target gene, targeting PirA Vp The gene sequence uses PrimerExpress3.0 software to design specific primers and TaqMan-MGB probes, the target product length is 60bp, the 5' end of the probe is labeled with the fluorescent group FAM, and the 3' end is labeled with the MGB group, primers and probe sequences for:
[0056] upstream primer PirA Vp -qF: 5'-CAAACGGAGGCGTCACAGA-3',
[0057] downstream primer PirA Vp -qR: 5′-GACCGACTTCCGGGATGAT-3′,
[0058] ProbePirA Vp -P: 5'-FAM-AGACAGCAAACATACACC-MGB-3'.
[0059] 2. DNA extraction and preparation of recombinant plasmid standards
[0060] (1)Vp AHPND Recovery and DNA extraction:
[0061] Take the frozen-thawed strain and inoculate it on TSA medium, incubate at 36±1°C for 18-24h, the...
Embodiment 2
[0083] sensitivity test
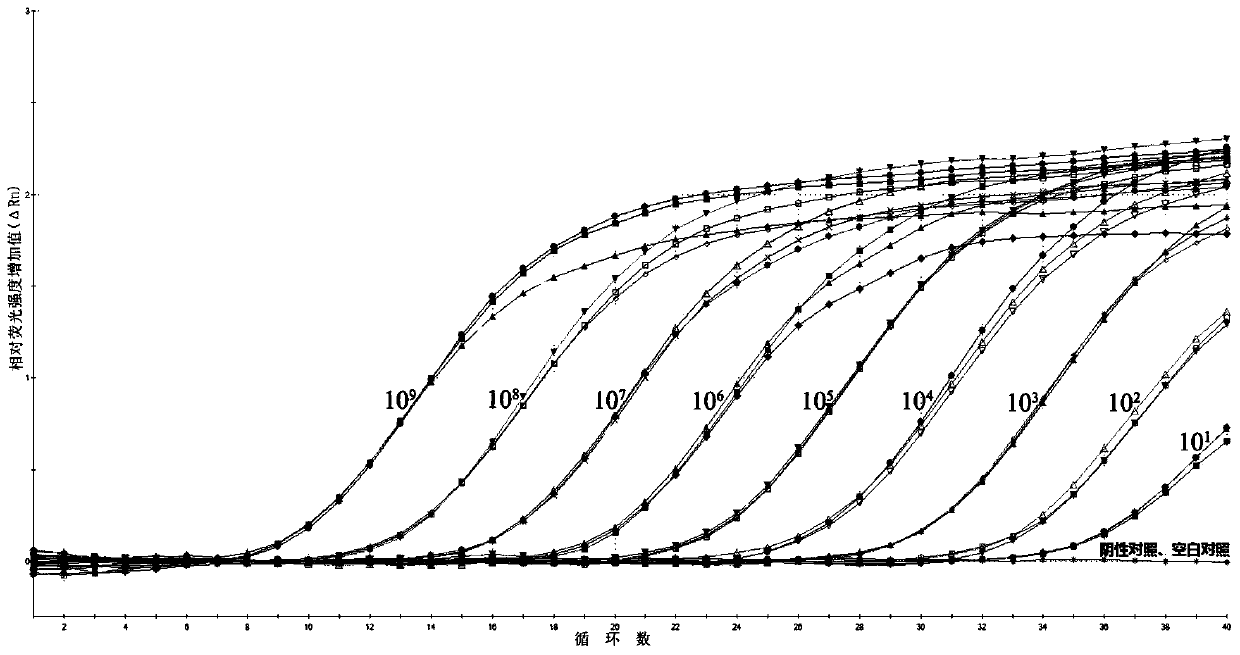
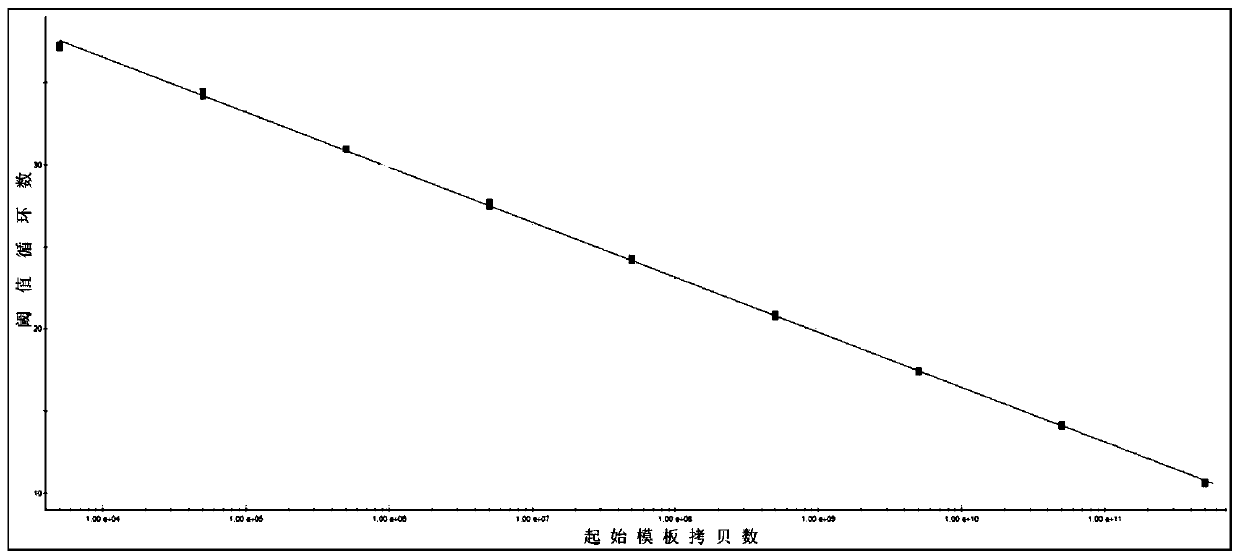
[0084] With 9 gradients (1.0×10 1 -1.0×10 9 copy / μL) standard DNA as a template, and use the optimized fluorescent quantitative PCR method to detect; use the data analysis software that comes with the instrument to analyze, and establish the logarithm and CT value (Y) of the initial template copy number (X) The standard curve and standard equation were used, and the quantitative range and sensitivity of the method were judged according to the amplification results of each standard DNA. Test results such as figure 2 (Standard (1.0×10 1 -1.0×10 9 copy / μL) fluorescence quantitative PCR amplification curve) and image 3 (Fluorescence quantitative PCR detection of Vp AHPND standard curve) shown.
[0085] Depend on figure 2 It can be seen that the 9 gradients (1.0×10 1 -1.0×10 9 copies / μL) of the standard DNA was amplified, and the results showed that: high-concentration standard (1.0×10 3 -1.0×10 9 copies / μL) of the amplification curves all s...
Embodiment 3
[0090] specificity test
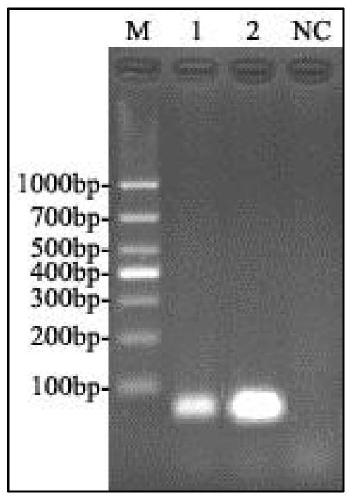
[0091] with Vp AHPND , Vibrio parahaemolyticus, Vibrio harveyi, Vibrio alginolyticus, Vibrio vulnificus, Vibrio riverine, WSSV, IHHNV, EHP and SHIV nucleic acid samples were used as templates, and the optimized fluorescent quantitative PCR method was used for detection and evaluation The specificity of the method, test results such as Figure 4 (Vp AHPND Specific detection results of fluorescent quantitative PCR).
[0092] Among them, Vibrio bacteria (Vp AHPND -BH170612 strain, Vibrio parahaemolyticus VP-FCG151118, Vibrio harveyi VH-QZ160809, Vibrio alginolyticus VA-QZ160809, Vibrio vulnificus VV-BH160905, Vibrio riverina VF-FCG170910) recovery and DNA extraction methods are : Streak inoculation of frozen and thawed Vibrio strains on TSA medium, culture at (36±1)°C for 18-24h, then select a single colony and inoculate on TSB medium, and continue to cultivate at (36±1)°C for 18-24h . Take 1.0-3.0 mL of the above bacterial solution, centrifuge at ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com