Method of assembling multiple gene fragments to py-2u vector via yeast and application
A technology of py-2u and gene fragments, applied in the field of bioengineering, can solve the problems of high mutation rate, time-consuming and laborious assembly of small fragments, difficulty in amplifying fragments above 10kb by polymerase chain reaction, etc., and achieve simple, convenient and high-speed cloning process Stirring and centrifugation, time-saving effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] refer to Figure 1-6 As shown, the method of using yeast to assemble multiple gene fragments into the py-2u vector specifically includes the following steps:
[0043]S1. Add a homologous sequence homologous to the py-2u vector at the two segments of the target gene to be cloned, with a length of 40-60bp;
[0044] S2. Divide the gene obtained in step S1 into a plurality of small fragments, which can usually be divided equally according to the length, and a separate enzyme cutting site is added to the two segments (this enzyme cutting site should not exist in the target gene, otherwise the enzyme will be used later) When a small fragment plasmid is digested with a cleavage site, the target gene containing the cleavage site will be cut). The restriction sites at both ends of each small fragment are the same restriction site, or different individual restriction sites.
[0045] S3. Each small fragment can be obtained by gene synthesis or by PCR amplification, and the blunt...
Embodiment 2
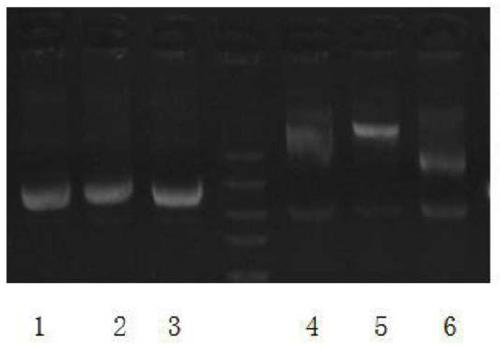
[0054] refer to figure 1 As shown, 3 vector plasmids were randomly selected, extracted after arabinose induction, and electrophoresed on the plasmids, see lanes 1, 2, and 3, and electrophoresed in lanes 4, 5, and 6 after extraction of plasmids in the uninduced control group, visible lanes The concentration of uninduced plasmid was lower, and the content of supercoiled plasmid was lower than that of induced plasmid. The values measured with a onedrop spectrophotometer are as follows:
[0055]
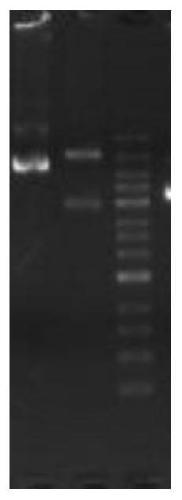
[0056] refer to figure 2 Shown is the final restriction map of the plasmid, digested with AscI and PmlI, the correct size is 9k / 5.2k. The time recording results of related operations are shown in the table below:
[0057]
[0058]
[0059] In this example, yeast can be used to assemble 8 gene fragments at a time, which greatly exceeds the upper limit of the fragments that can be assembled by traditional assembly methods (2-3 gene fragments, even if it is Gibson, the succes...
Embodiment 3
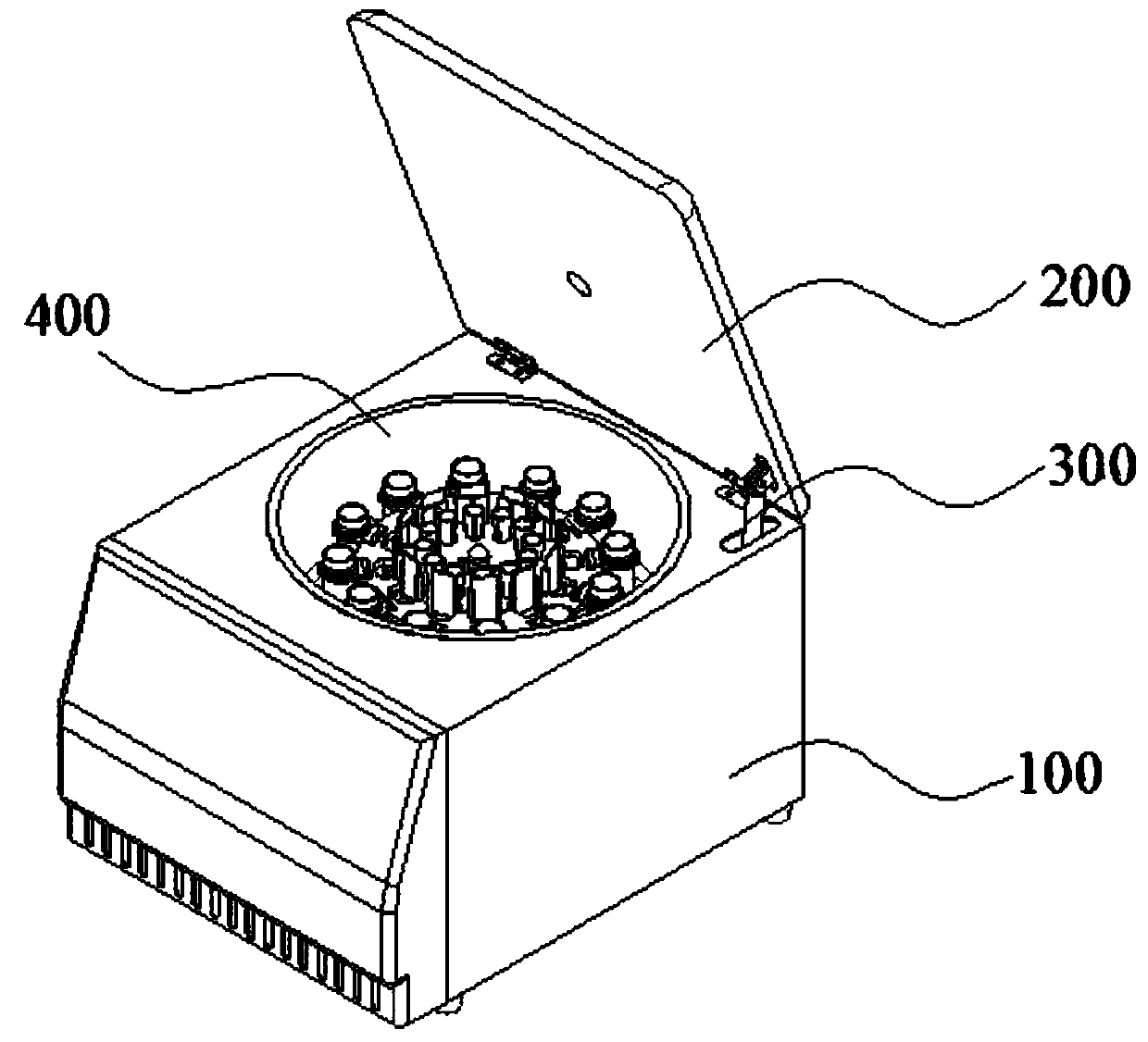
[0063] refer to Figure 3-6 , This embodiment provides a constant temperature incubation device, which can accommodate, centrifugally stir, and heat various test tubes and glass slides of different specifications and sizes. Specifically, the constant temperature incubation device includes a thermal insulation shell 100, an airtight door 200, and accommodates a centrifugal mechanism. The interior of the thermal insulation shell 100 has a cylindrical cavity, and the airtight door 200 is used to accommodate the opening and closing of the centrifugal mechanism. The top of the thermal insulation shell 100 is hinged, and the airtight door 200 is connected with the bottom of the thermal insulation shell 100 with a cylinder telescopic rod 300 .
[0064] The centrifuge accommodating mechanism is arranged in the cylindrical cavity inside the thermal insulation shell 100, including: a shock-absorbing disc 400, a shock-absorbing installation mechanism, and a multi-test tube accommodating ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com