Application of histone H2B mono-ubiquitination for identifying homologous recombination repair defects
A homologous recombination, monoubiquitin technology, applied in biochemical equipment and methods, microbial determination/inspection, instruments, etc., can solve the problem of difficulty in judging BRCA1/BRCA2 function loss, inability to guide the use of targeted drugs, homologous recombination The clinical diagnosis of defects and other problems are complicated, so as to achieve the advantages of good clinical economics and facilitate clinical operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
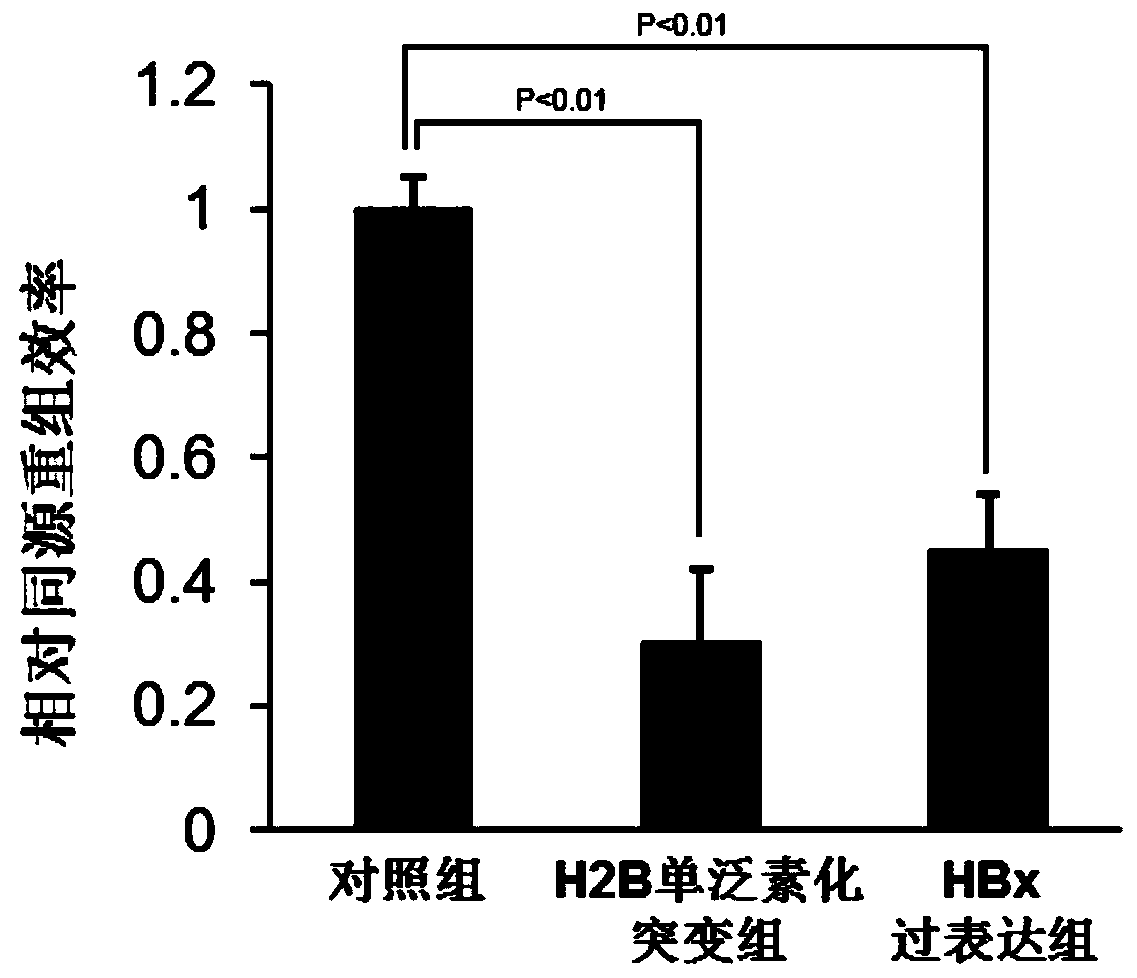
[0043] Example 1. H2B monoubiquitination mutant reduces homologous recombination repair efficiency
[0044] Experimental method: Inoculate HL-L02 cells into a 6cm culture dish, culture at 37°C until the cell coverage reaches 50-70%, transfect pLVX-Green-H2BK120RK125R(2KR), pLVX-Green-HBx and control plasmid pLVX-Green , after continuing to culture for 24 hours, transfect the I-SCE-I-HR plasmid, and use the I-SCE-I system to detect the efficiency of homologous recombination in vivo: collect the cells after 24 hours, and pass the High Pure PCR Template Preparation kit (Roche, 11796828001) Genomic DNA was extracted from cells, and primers for detecting HR efficiency were designed (primer sequence F: TGACCACCCCTGACCTACG; R: CACCTTGATGCCGTTCTTCTGC), and real-time fluorescent quantitative PCR was used to identify the effect of H2B monoubiquitin mutants on homologous recombination in vivo.
[0045] Experimental results: This experiment uses the I-SceI-GFP system (Development of Novel...
Embodiment 2
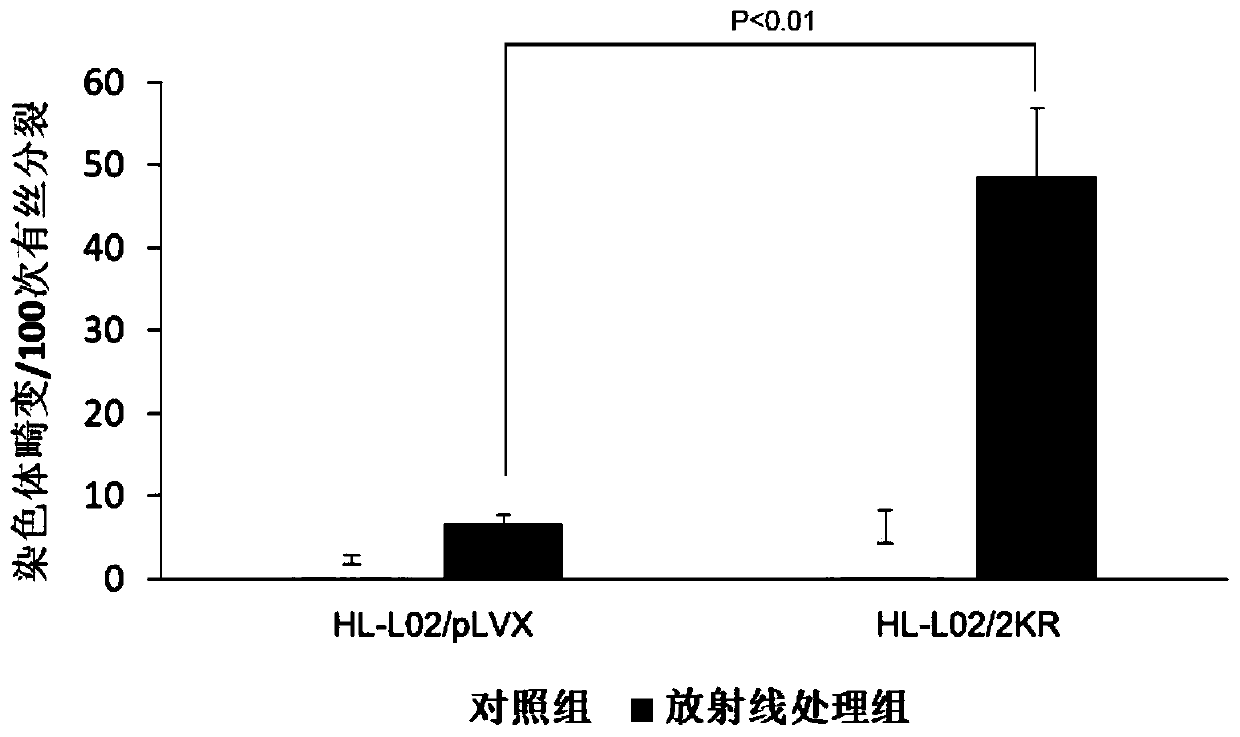
[0047] Example 2. H2B monoubiquitination mutants cause massive chromosomal breaks
[0048]Experimental method: The H2B monoubiquitination mutant cell line HL-L02 / 2KR and the control cell HL-L02 / pLVX were constructed using a lentiviral infection system. The constructed cells were inoculated into two 6cm-diameter culture dishes and cultured overnight. Eight hours after X-ray treatment (dose of 4Gy), colchicine (final concentration 200ng / ml) was added for 1.5 hours. Trypsinized, washed with PBS. Remove the supernatant and add 250 μl PBS to resuspend the cells. Add 6ml of 37°C preheated 75mM KCl dropwise. Incubate at 37°C for 25 minutes. Add 200 μl fixative (methanol: glacial acetic acid = 3:1) dropwise, mix gently, and centrifuge at 1000 rpm for 10 minutes. Add 5ml of fixative, mix well, and let stand at 4°C for 20 minutes. Centrifuge at 1000rpm for 10 minutes, and resuspend the cells in 50ul of fixative. Repeatedly fix the cells 3 times, and finally add 500 μl of fixative...
Embodiment 3
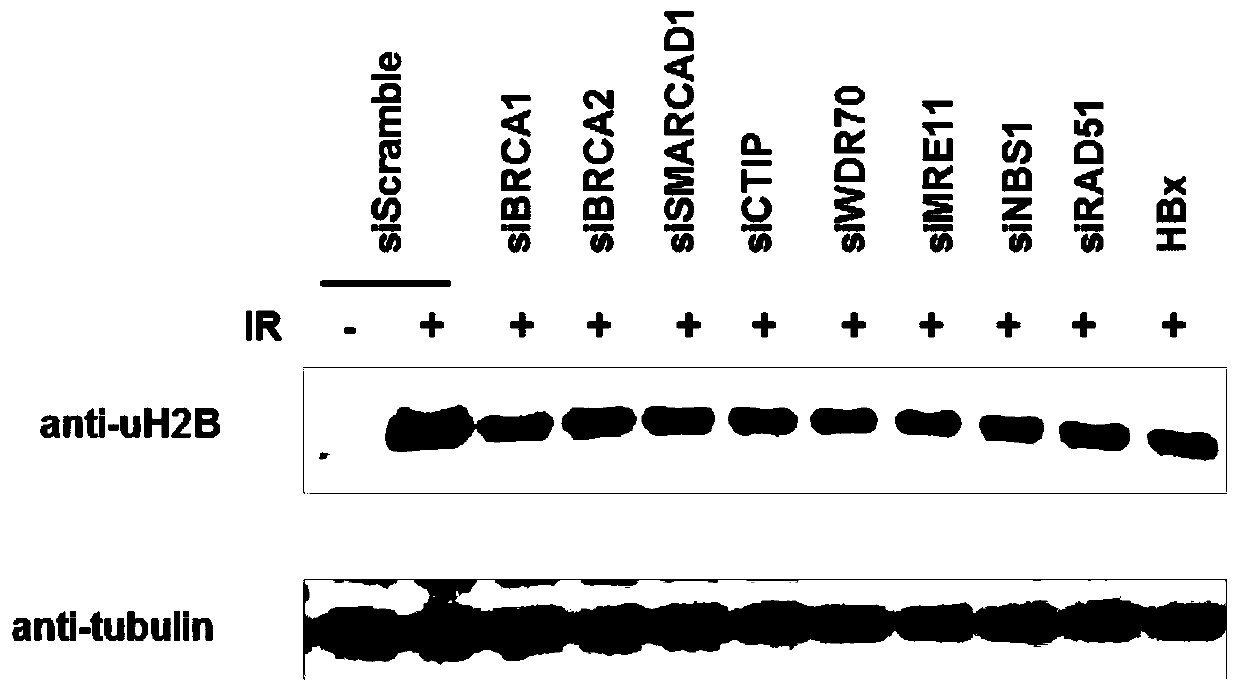
[0051] Example 3. Deficiency in homologous recombination repair leads to reduced H2B monoubiquitination levels
[0052] Experimental method: Small interfering RNA (siRNA) was used to silence homologous recombination repair genes such as BRCA1, BRCA2, WDR70, CTIP, MRE11, NBS1 and RAD51 in HL-L02 cells, random siRNA (Scramble) was used as the experimental control, and pLVX- Green-HBx plasmid. siRNA or HBx plasmid was transfected and silenced for 48 hours, then irradiated with ion rays (dose 10Gy), 4 hours later the total protein was extracted, and the H2B monoubiquitination level in the cells was detected by immunoblotting.
[0053] Experimental results: BRCA1, BRCA2, WDR70, CTIP,
[0054] The silencing of homologous recombination repair genes such as SMARCAD1, MRE11, NBS1 and RAD51 and the expression of the oncogene HBx all resulted in a decrease in the level of H2B monoubiquitination.
[0055] Conclusion: The results indicate that the functional abnormality of key components...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com