Mutant glyphosate degrading enzyme and cloning, expression and application thereof
A glyphosate and enzyme-degrading technology, applied in the field of molecular biology, can solve problems such as food threats, glyphosate activity reduction, and limited application development, achieving efficient decomposition, large application potential, and the effect of solving residual problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Study on site-directed mutation of the 63rd amino acid of glyphosate degrading enzyme
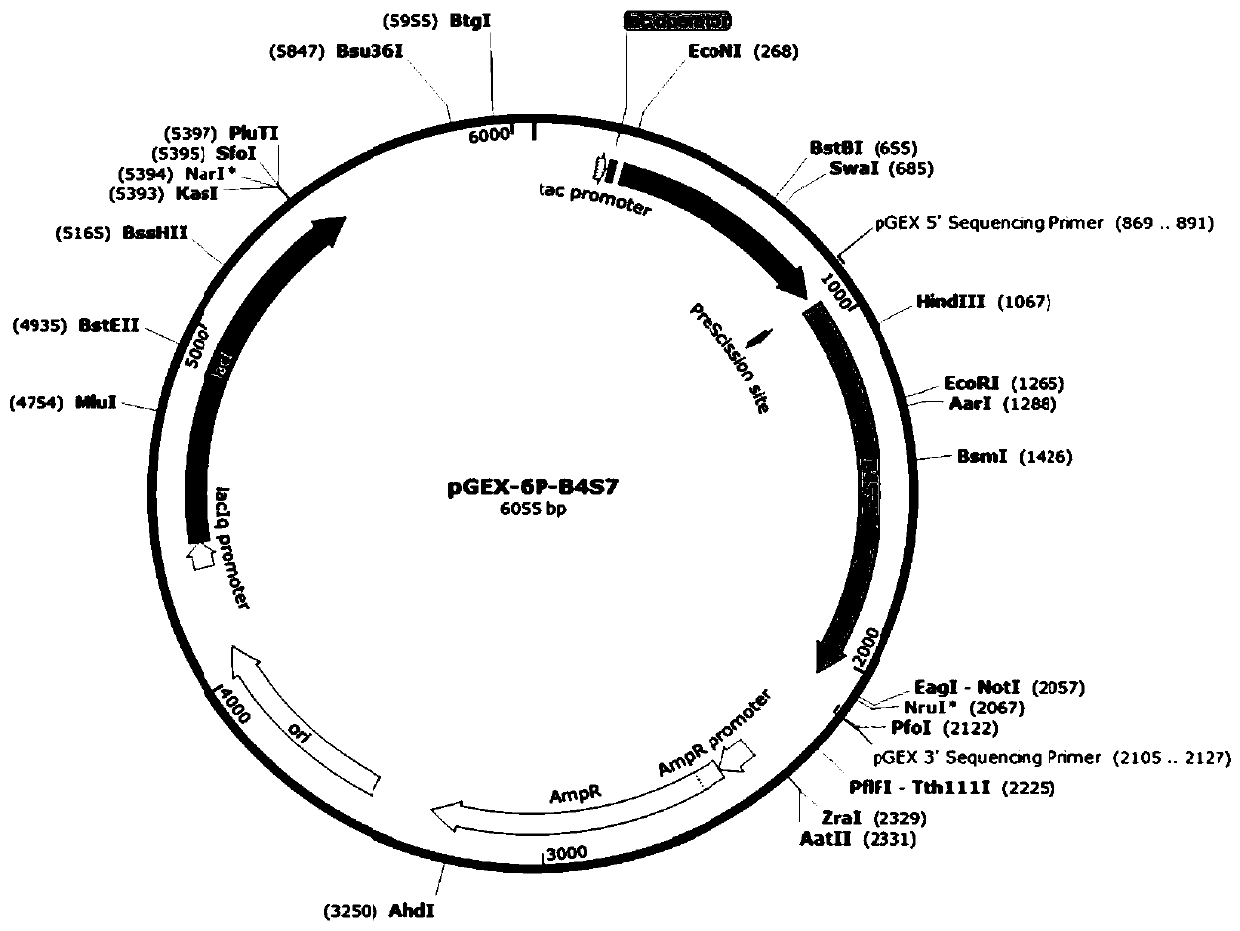
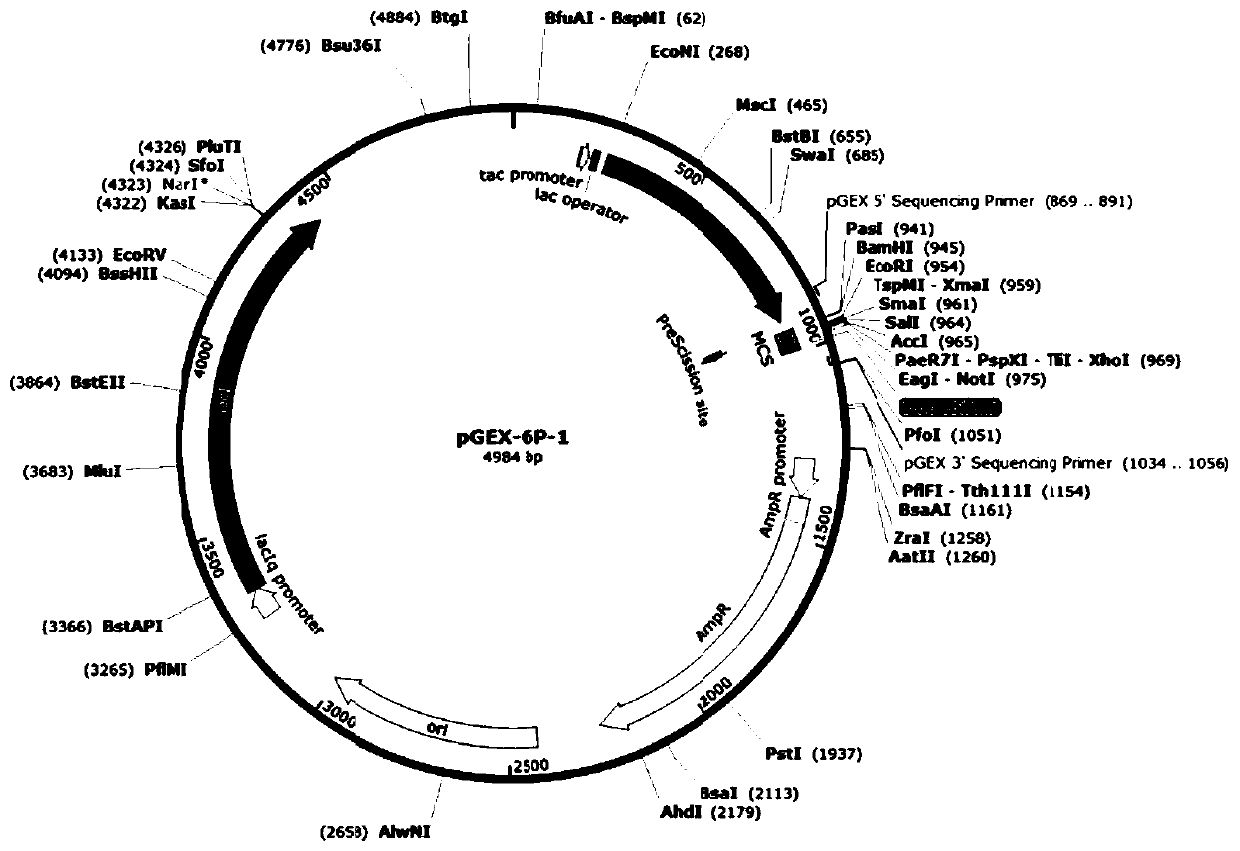
[0028] With the recombinant plasmid pGEX-6p-B4S7 (see the attached figure 1 (shown) is the template (pre-laboratory construction, reference Int J Biol Macromol, 2015, 79: 965-970), using the designed circular expansion primer (see Table 1 below), oligonucleotide-mediated The PCR site-directed mutagenesis method (guided according to the QuikChange Site-directedMutagenesis kit kit of Stratagene Company) introduced a mutation at the 63rd amino acid residue site of glyphosate degrading enzyme, and its coding gene was passed through pGEX-6p-1 (purchased From GE Healthcare in the United States, its plasmid map is attached figure 2 Shown) on the plasmid vector and recombined into pGEX-6p-F63X plasmid.
[0029] The specific PCR reaction conditions are: pre-denaturation at 97°C for 2 min; denaturation at 95°C for 20 sec; annealing at 54°C for 30 sec; extension at 72°C for 2 min a...
Embodiment 2
[0033] Example 2 Glyphosate degrading enzyme mutant protein expression and purification
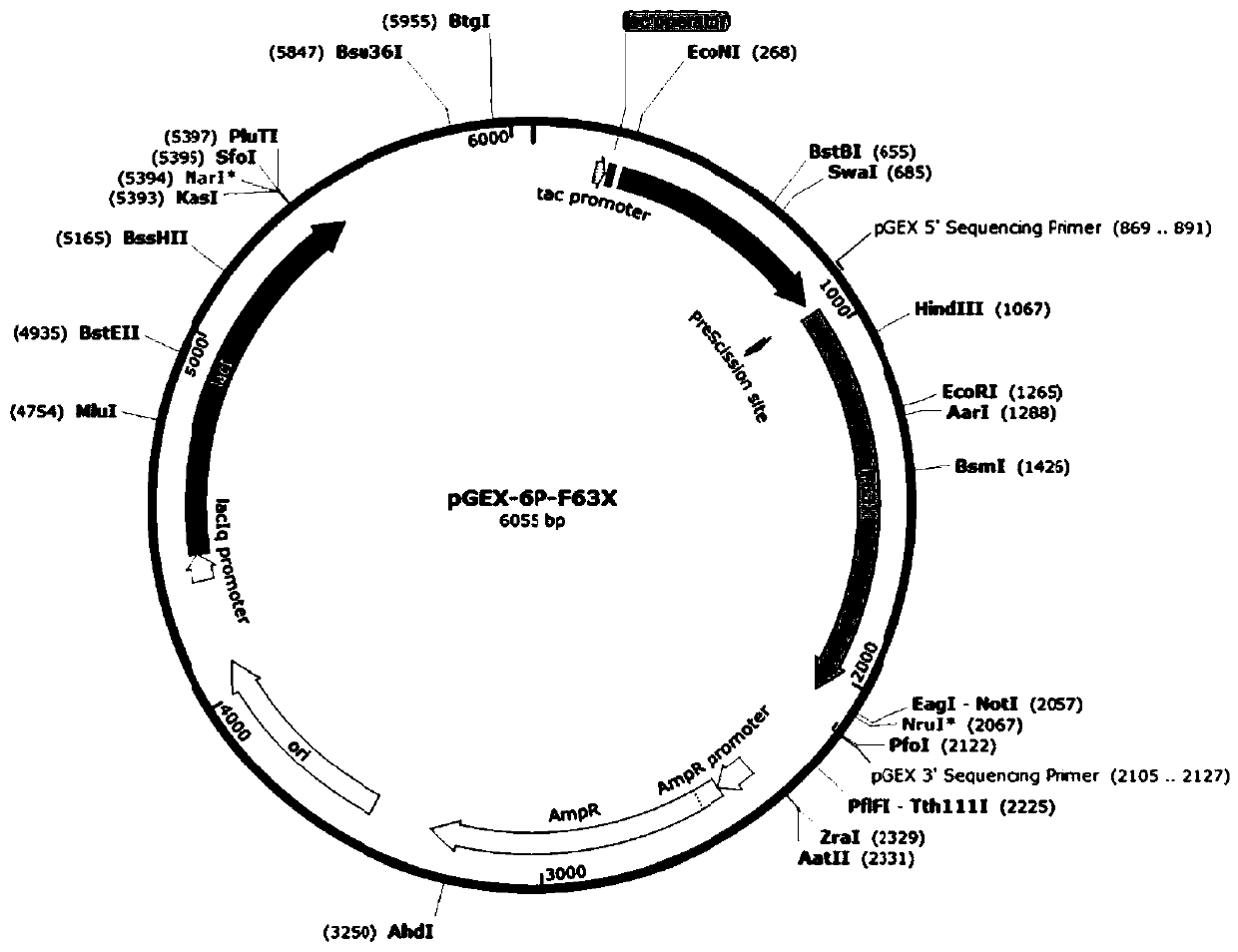
[0034] The recombinant plasmid pGEX-6p-F63X containing the mutant F63X gene (see attached image 3 shown) were mixed with E.coli BL21(DE3) competent cells, transformed by electric shock, spread on ampicillin-resistant plates (ampicillin concentration 100 μg / mL), and cultured overnight at 37°C until a single colony visible to the naked eye grew. Pick a single clone and inoculate it into 20 mL of liquid LB medium (containing 100 μg / mL of ampicillin), and culture overnight at 37° C. with shaking at 200 rpm. Transfer 1% of the bacterial solution to 2L of fresh liquid LB medium (containing ampicillin 100 μg / mL), shake culture at 37°C and 200rpm until the OD600 is 0.6, add the inducer IPTG with a final concentration of 0.1mM, and store at 22°C, 160rpm induction culture for 12h to express the protein. The bacteria were collected by centrifugation, washed twice with 50mM sodium pyrophosphate bu...
Embodiment 3
[0037] Embodiment 3 glyphosate degrading enzyme mutant enzymatic kinetics assay
[0038] In the present embodiment, the enzyme activity assay of glyphosate degrading enzyme and its mutant adopts the horseradish peroxidase coupling method (Job, Marcone et al.2002), by measuring the H released by the reaction 2 o 2 The amount reflects the enzyme activity. Using o-dianisidine as a chromogenic substrate, glyphosate is oxidized and decomposed by glyphosate-degrading enzymes to produce H 2 o 2 Oxygen is released by horseradish peroxidase to oxidize o-dianisidine to show orange red, with an absorption peak at 450nm, and measure OD 450 The value was entered into the standard curve to calculate the enzyme activity.
[0039] (1) Enzyme activity assay method
[0040] Convert 1 μmoL of substrate (containing glycine or oxygen) or generate 1 μmoL of product H at 25°C and optimal pH 2 o 2 The required enzyme amount of glyphosate-degrading enzyme is defined as one enzyme activity unit ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| catalytic efficiency | aaaaa | aaaaa |
| catalytic efficiency | aaaaa | aaaaa |
| catalytic efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com