Method for transforming bacillus subtilis FMMs and application thereof
A Bacillus subtilis, domain technology, applied in the field of transforming Bacillus subtilis FMMs, can solve problems such as lysis and severe cells, and achieve the effects of simple construction method, good application prospects, and easy use.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
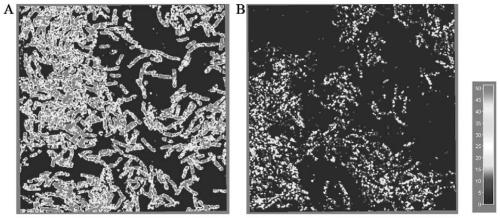
[0036] The transformation and verification of the FMMs of embodiment 1 Bacillus subtilis
[0037] On the basis of Bacillus subtilis BSGN6, the yisp homologous recombination integration expression frame is integrated at the tcyp or yhcm site on the Bacillus subtilis BSGN6 genome. Use integration site tcrp sequence (length 1kb), spectinomycin resistance gene spc sequence (nucleotide sequence shown in SEQ ID NO.5, including lox site), yisp gene sequence, yhcM downstream sequence (length 1kb) Build the integration box. Through homologous recombination, the integration frame obtained above was integrated into the genome of Bacillus subtilis BSGN6. Through spectinomycin resistance plate screening, colony PCR verification, and sequencing, it was confirmed that the integration was successful and the recombinant Bacillus subtilis overexpressing yisp was obtained. The SPFH gene fragment was inserted into the multiple cloning site of the pP43NMK plasmid using Gibson seamless assembly t...
Embodiment 2
[0039] Embodiment 2 analyzes the impact of FMMs remodeling on the growth of Bacillus subtilis cells
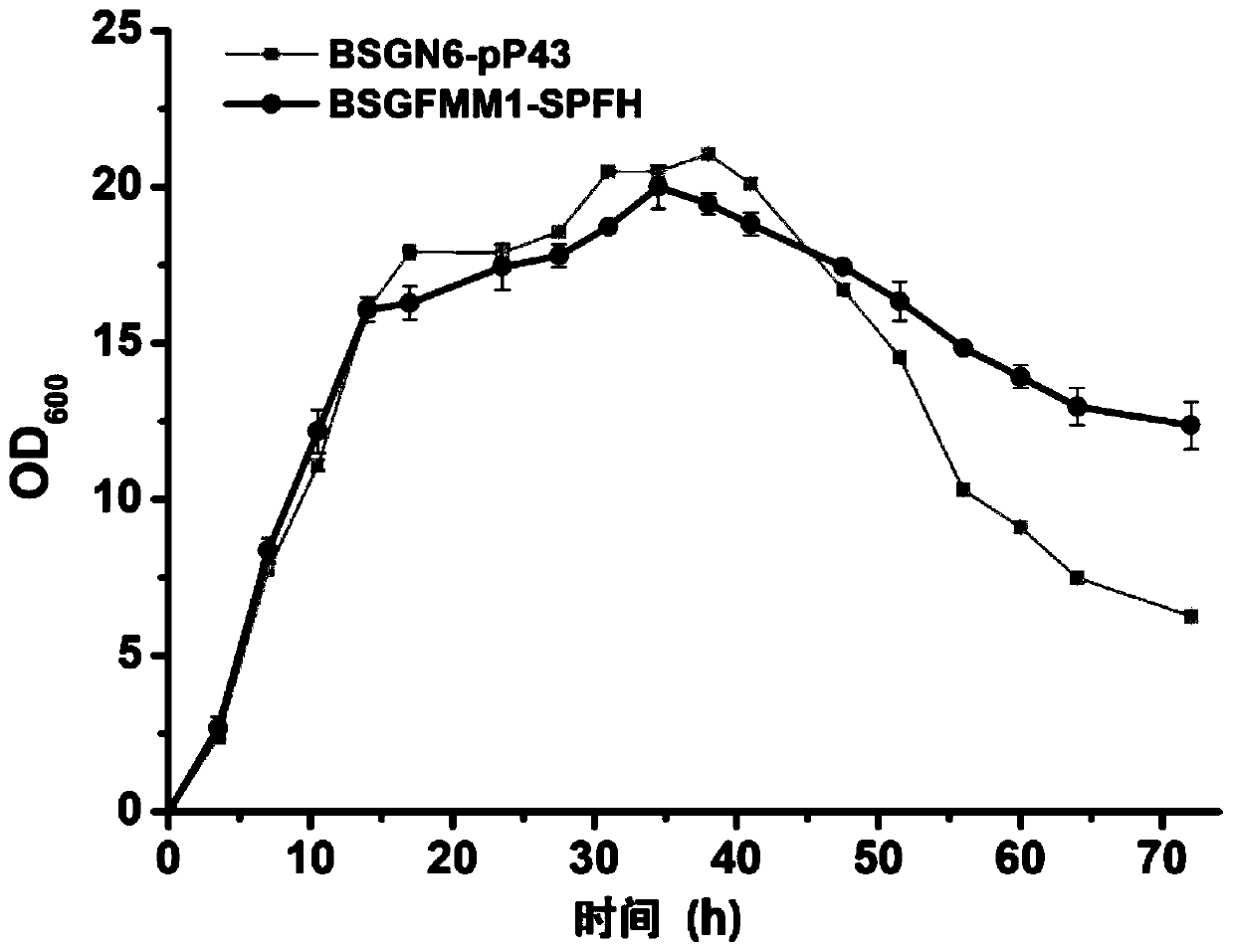
[0040] Cultivate the transformant of BSGN6-pP43 and BSGFMM1 recombinant Bacillus subtilis in seed medium (10g / L peptone, 5g / L yeast powder and 10g / L sodium chloride) for 8-10h, then follow the inoculum size of 2-5% Transfer to fermentation medium (40g / L glucose, 6g / L peptone, 12g / L yeast powder, 6g / L ammonium sulfate, 12.5g / L dipotassium hydrogen phosphate, 2.5g / L potassium dihydrogen phosphate, 5g / L carbonic acid Calcium 10ml / L trace elements), cultured in a 250mL shake flask at 37°C and 220rpm for 72h.
[0041] Use UV-visible spectrophotometer to regularly measure the absorbance value OD of fermentation broth 600 To characterize cell growth, such as figure 2 Therefore, the OD of BSGN6-pP43 and BSGFMM1 strains within 45h 600 There was no significant difference in the value, after 45h, the OD of BSGFMM1 600 The value was significantly higher than that of BSGN6-pP43 strain...
Embodiment 3
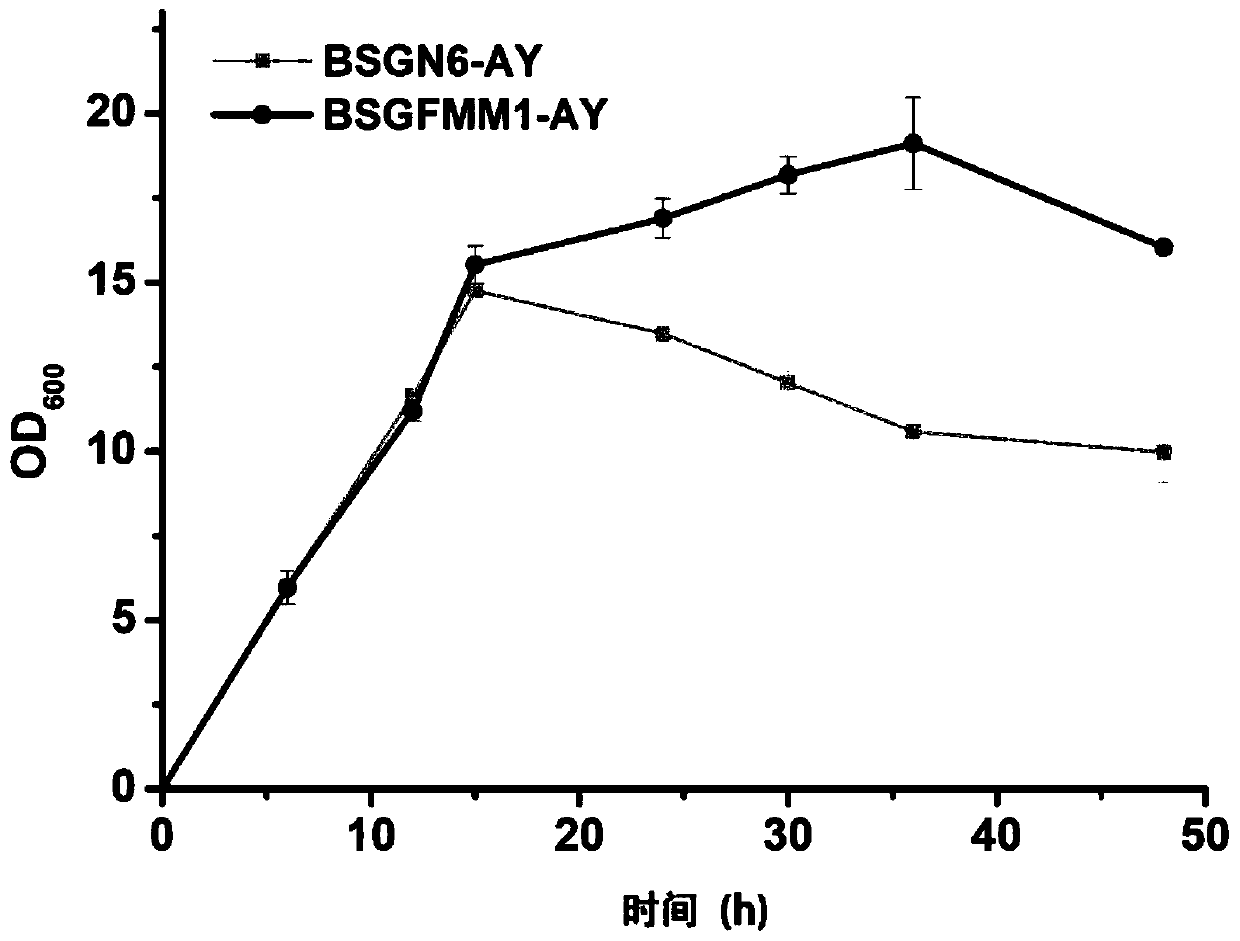
[0042] Example 3 Construction of recombinant Bacillus subtilis BSGFMM1-AY
[0043] On the basis of Bacillus subtilis BSGFMM1, the fusion gene expression cassette of SPFH and yqaB was integrated at the hprK site of Bacillus subtilis BSGFMM1 genome. Using the integration site hprK sequence (length 1kb), the chloramphenicol resistance gene CmR sequence (the nucleotide sequence is shown in SEQ ID NO.6, including the lox site), the C-terminal SPFH gene sequence with GS linker ( 0.68kb), yqaB gene sequence (0.57kb), nagR gene and its downstream gene sequence (length 1kb) to construct the SPFH-yqaB fusion gene integration frame, wherein, SPFH and yqaB are fusion expressions, and the linker nucleotides connected between the two The acid sequence is shown in SEQID NO.4. The integration frame obtained above was integrated into the genome of Bacillus subtilis BSGFMM1 through homologous recombination. Through chloramphenicol resistance plate screening, colony PCR verification, and seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com