Portunus trituberculatus C-type lectin PtCLec2 gene and encoded protein thereof and application of encoded protein
A technology of Portunus trituberculatus and gene encoding, which is applied in the field of molecular biology and can solve problems such as unclear influence of gene function
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
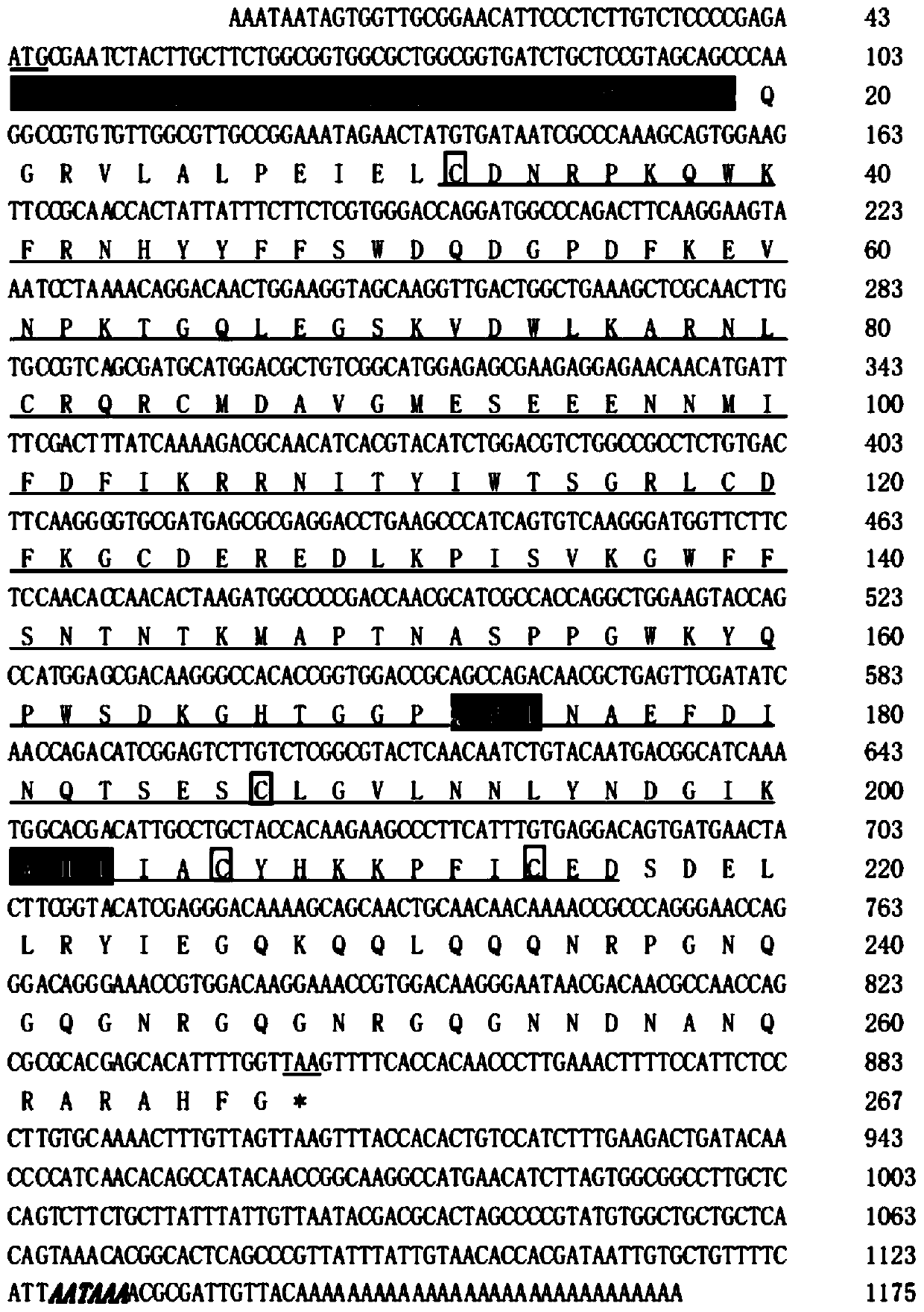
[0029] The C-type lectin PtCLec2 gene of Portunus trituratus is the base sequence shown in SEQ ID No.1.
[0030] see figure 1 , the sequence listing SEQ ID No.1 is:
[0031]AAATAATAGTGGTTGCGGAACATTCCCTCTTGTCTCCCCGAGAATGCGAATCTACTTGCTTCTGGCGGTGGCGCTGGCGGTGATCTGCTCCGTAGCAGCCCAAGGCCGTGTGTTGGCGTTGCCGGAAATAGAACTATGTGATAATCGCCCAAAGCAGTGGAAGTTCCGCAACCACTATTATTTCTTCTCGTGGGACCAGGATGGCCCAGACTTCAAGGAAGTAAATCCTAAAACAGGACAACTGGAAGGTAGCAAGGTTGACTGGCTGAAAGCTCGCAACTTGTGCCGTCAGCGATGCATGGACGCTGTCGGCATGGAGAGCGAAGAGGAGAACAACATGATTTTCGACTTTATCAAAAGACGCAACATCACGTACATCTGGACGTCTGGCCGCCTCTGTGACTTCAAGGGGTGCGATGAGCGCGAGGACCTGAAGCCCATCAGTGTCAAGGGATGGTTCTTCTCCAACACCAACACTAAGATGGCCCCGACCAACGCATCGCCACCAGGCTGGAAGTACCAGCCATGGAGCGACAAGGGCCACACCGGTGGACCGCAGCCAGACAACGCTGAGTTCGATATCAACCAGACATCGGAGTCTTGTCTCGGCGTACTCAACAATCTGTACAATGACGGCATCAAATGGCACGACATTGCCTGCTACCACAAGAAGCCCTTCATTTGTGAGGACAGTGATGAACTACTTCGGTACATCGAGGGACAAAAGCAGCAACTGCAACAACAAAACCGCCCAGGGAACCAGGGACAGGGAAACCGTGGACAAGGAAACCGTGGACAAGGGAATAACGACAACGCCAA...
Embodiment 2
[0064] The base sequence of the C-type lectin of Portunus trituratus is described in SEQ ID No. 1 of the sequence listing, and the amino acid sequence is described in SEQ ID No. 2 of the sequence listing.
[0065] Sequence Listing SEQ ID No.2 is:
[0066] MRIYLLLAVALAVICSVAAQGRVLALPEIELCDNRPKQWKFRNHYYFFSWDQDGPDFKEVNPKTGQLEGSKVDWLKARNLCRQRCMDAVGMESEEENNMIFDFIKRRNITYIWTSGRLCDFKGCDEREDLKPISVKGWFFSNTNTKMAPTNASPPGWKYQPWSDKGHTGGPQPDNAEFDINQTSESCLGVLNNLYNDGIKWHDIACYGRGHKKPFICEDSDQELLYARANHRPGQFGRGRG
[0067] It has a complete coding protein containing 267 amino acids, the coding sequence has a signal peptide (1-19), the predicted molecular weight is 30.65kDa, and the isoelectric point is 7.53. The mature peptide contains 248 amino acids with a typical CRD domain (32-215), which contains 4 cysteine residues capable of forming two disulfide bonds. The CRD domain lacks the typical vertebrate and invertebrate mannose-recognizing motif EPN (Glu-Pro-Asn) but contains galactose-recognizi...
Embodiment 3
[0071] 1. In vitro antibacterial test of C-type lectin PtCLec2 recombinant protein of Portunus trituratus
[0072] Culture and preparation of microorganisms: Vibrio alginolyticus was cultured with TSB medium at 28°C, Pseudomonas aeruginosa was cultured with TSB medium at 37°C, Staphylococcus aureus was cultured with LB medium at 37°C, and Micrococcus luteus was cultured with LB. The base was cultured at 37°C, Pichia pastoris was cultured with YPD medium at 28°C, and the above strains were cultured with a shaker at 220 rpm / min so that the bacterial concentration reached the logarithmic growth phase, and were diluted with 50 mM Tris-HCl (pH=8.0) buffer solution. Bacteria, so that the number of colonies per milliliter of bacterial solution is about 1 × 10 3 indivual.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com