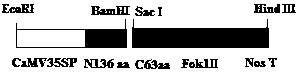TALE nuclease simplified framework construction for plant gene fixed-site shearing
A nuclease and plant technology, applied in plant genetic improvement, plant products, genetic engineering, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1: Chemical Synthesis of the Streamlined Backbone Coding Sequences for Targeted Editing of Plant Gene TALENs
[0019] The simplified framework of TALENs was synthesized by gene synthesis method (Nucleic Acids Research, 2004, 32, e98). The designed primers are:
[0020] SplantTALENs-1 ATGGACCCAATCCGTTCTCGTACTCCATCTCCTGCACGTGAACTGCTGCCTGGTCCACAA (shown in SEQ ID NO.2)
[0021] SplantTALENs-2 GTGCACCACCACGATCAGCAGTAGGCTGGACCCTGTCAGGTTGTGGACCAGGCAGCAGTT (shown in SEQ ID NO.3)
[0022] SplantTALENs-3 TGCTGATCGTGGTGGTGCACCACCTGCTGGTGGTCCACTGGACGGTCTTCCTGCTCGTCG (shown in SEQ ID NO.4)
[0023] SplantTALENs-4 TGGTGCAGGTGGAGATGGCAGCCGAGTCCGAGACATAGTACGACGAGCAGGAAGACCGTC (shown in SEQ ID NO.5)
[0024] SplantTALENs-5 TGCCATCTCCACCTGCACCATCTCCTGCATTCTCTGCTGGTTCCTTTCTCCGACCTGCTGC (shown in SEQ ID NO.6)
[0025] SplantTALENs-6 TCAAGAAGGGAGGTGTCAAGAAGGGATGGATCGAACTGACGCAGCAGGTCGGAGAAGGAA (shown in SEQ ID NO.7)
[0026] SplantTALENs-7 CTTGACACCCTCCCTTCTTGACTCGATGCCTGCTGTT...
Embodiment 2
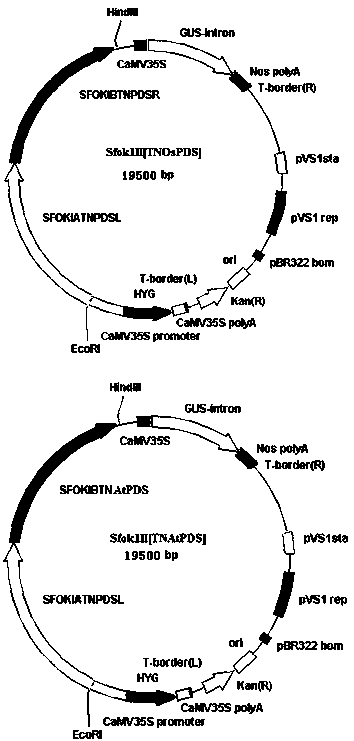
[0052] Example 2: Chemical Construction of Plant Expression Elements for Simplified Skeletons of Plant Gene TALENs
[0053] The simplified skeleton of plant TALENs The promoter of SplantTALENs is CaMV35S+Omega [Omega is the translation enhancer from TMV virus], the terminator is Nos, and the N-terminal and C-terminal sequences required by TALEN are added to the N-terminal of the FokI cleavage domain ( Including the N-terminal 163 amino acid sequence and the C-terminal 63aa skeleton), a small fragment of DNA sequence with BamHI and SacI restriction endonuclease sites at both ends was introduced between the N-terminal and C-terminal sequences for insertion The above constructed target site binding functional modules. Both ends of SplantTALENs have EcoRI and HindIII cutting sites, which facilitate the cloning and identification of plant expression units
[0054] The CaMV35S+Omega and Nos terminators were amplified by PCR from a conventional vector, and then the promoters, the re...
Embodiment 3
[0062] Example 3: The streamlined backbone of plant gene TALENs is used for plant gene splicing
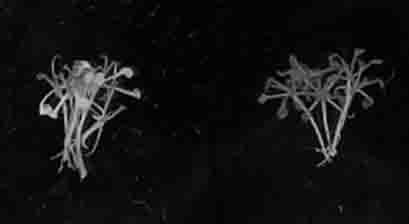
[0063]Phytoene desaturase (PDS) is a key enzyme in the carotenoid synthesis pathway. The inactivation of PDS gene function will lead to the ineffective synthesis of carotenoids, and the chlorophyll will be destroyed under light conditions, making Tissue that should be green turns white, a phenomenon known as photobleaching, which provides a visually apparent phenotypic change for loss of gene function.
[0064] In order to verify the application effect of TALENs streamlined framework in plant gene splicing. In this study, 15 bases of the target site were selected according to the genomic DNA sequence of the rice OsPDS1 gene and the Arabidopsis AtPDS1 gene, and the uniqueness of the target site in the rice genome was guaranteed by Blast analysis with the rice genome. According to the recognition of base T by NG, the recognition of base C by HD, the recognition of base A by NI, the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com