Recombinant phospholipase D and application thereof for synthesizing phosphatidylserine or other phospholipids
A phospholipase and phospholipid technology, applied in the field of functional enzyme screening, can solve the problems of overnight incubation, unfavorable PLD production or application, low yield, etc., and achieve the effect of improving transesterification activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
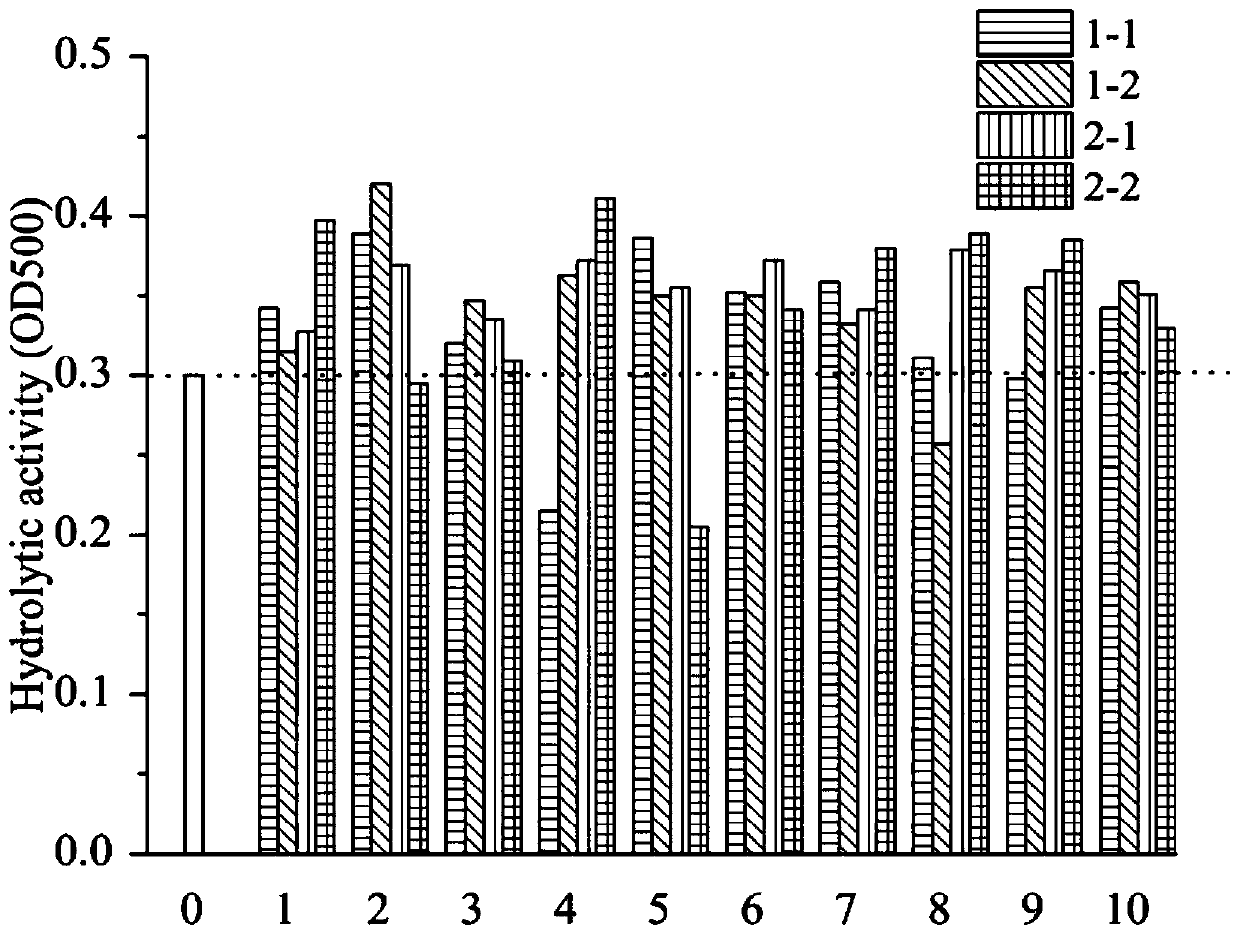
[0027] Embodiment 1: Screening and preparation of recombinant PLD
[0028] 1. PCR amplification of phospholipase D fragment
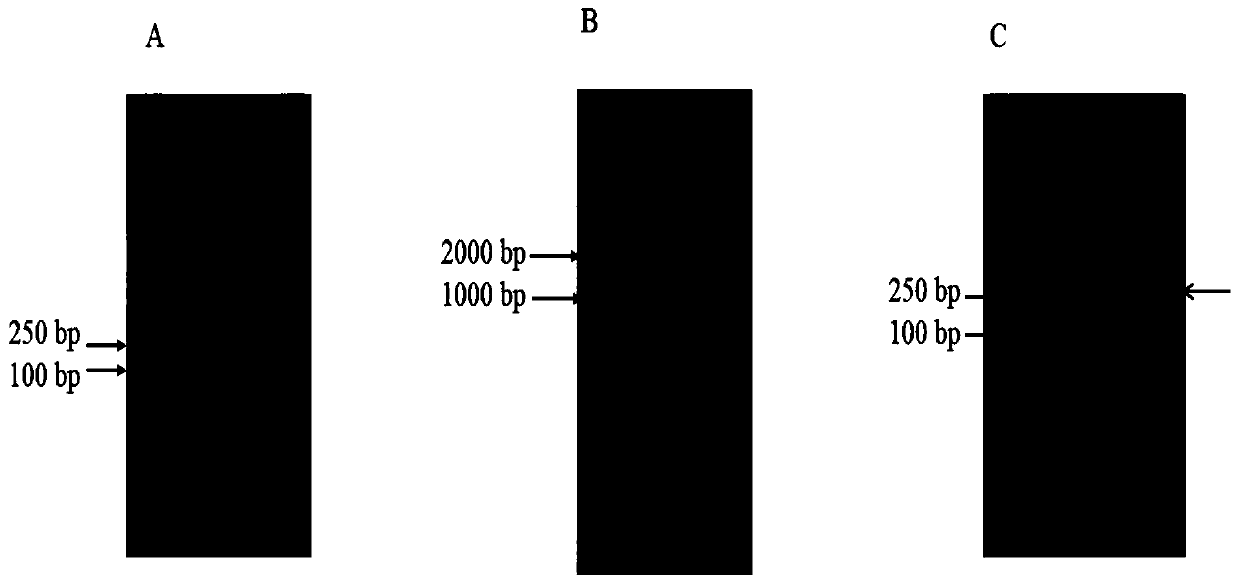
[0029] Firstly, the two PLD proteins obtained by the applicant were amplified by PCR method, namely S.griseofuscusstrain PLDsg / G213S and S.roseus, PLDsr, with a size of about 1500-1600bp. The amino acid sequence of PLDsg / G213S is SEQ ID NO :3, the sequence of the coding gene is SEQ ID NO:4; the amino acid sequence of S.roseus, PLDsr is SEQ ID NO:5, and the sequence of the coding gene is SEQ ID NO:6. Then the PCR product was recovered, and the concentrations of the two fragments were measured with a micro-nucleic acid analyzer. Get the same amount of PCR product solutions that have been amplified again and mix them with enzyme DNsae I to cut into fragments, and the enzyme-cut products are detected by agarose gel electrophoresis ( figure 1 A). According to the materials and methods, 0.3 μL DNase I (diluted to 1U / μL, diluted immediately after use) at 15...
Embodiment 2
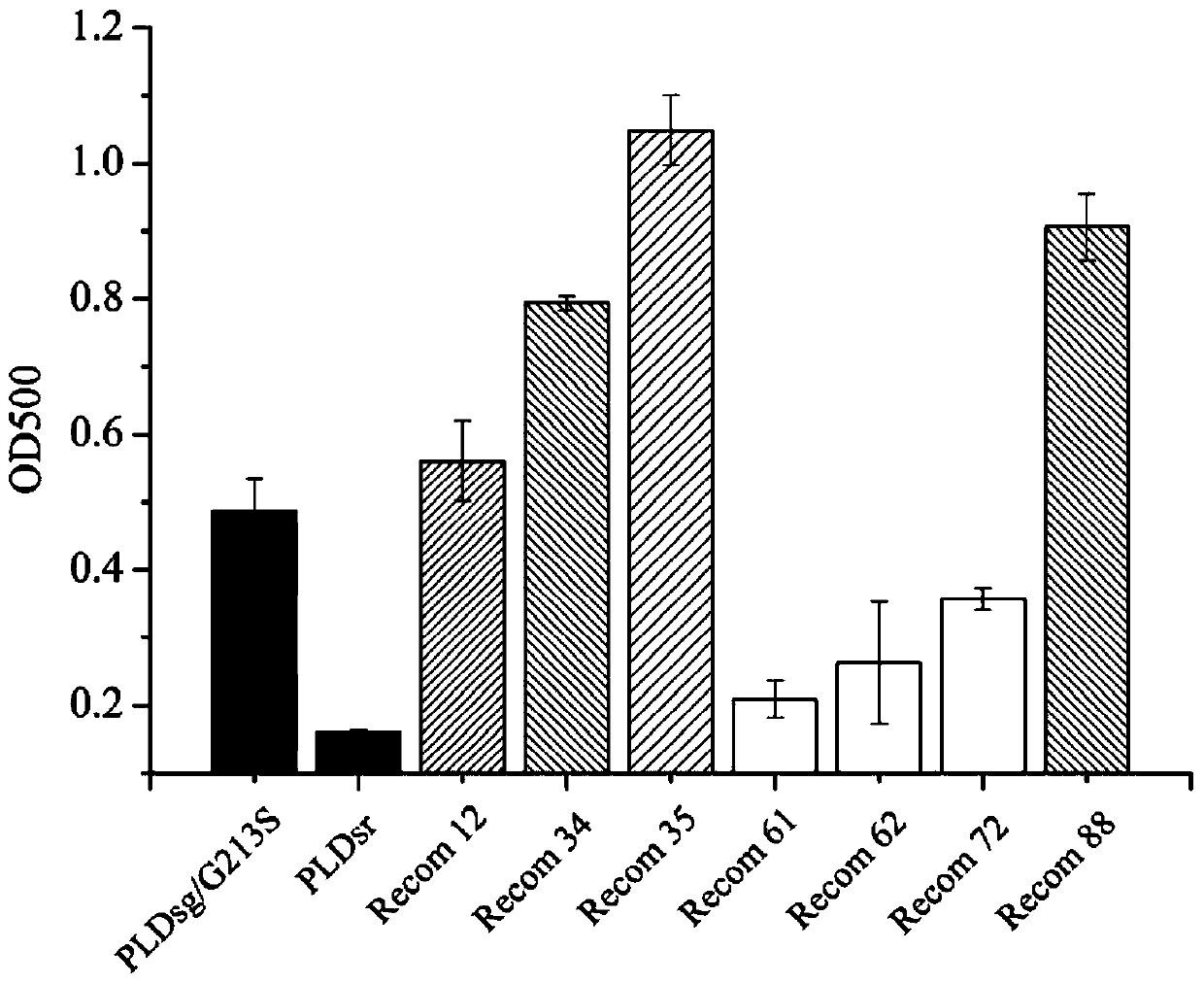
[0039] Embodiment 2: Recombinant PLDs enzyme synthesis PS and DHA-PS
[0040] In order to verify the ability of the recombinant PLDs enzyme of the present invention and the parent enzyme to synthesize PS, a two-phase reaction system was used to synthesize PS with phosphatidylcholine PC and L-serine as substrates. The parent enzymes PLDsr and PLDsg / G213S were used as controls, and Reom-12, Recom-34 and Recom-35 were used as experimental groups, and the same weight of enzyme powder, 1M L-serine and 50mM CaCl 2 Dissolve in 1mL HAc-NaAc buffer (20mM, pH 6.0), and the organic phase contains PC (dissolved in anhydrous ether, 20mg / mL). The enzymatic reaction time was optimized, and after 6 hours of reaction, the organic phase was collected by centrifugation, and the reaction results were detected by thin-layer chromatography and HPLC.
[0041] The result is as Figure 4 As shown in a, the first lane from left to right is the substrate phosphatidylcholine PC, the second and third la...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com