ssDNA aptamer for specifically identifying metronidazole and application thereof
A metronidazole and aptamer technology, applied in the field of ssDNA aptamers, can solve the problems of high price, non-immunogenicity, production and application limitations, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Embodiment 1: Construction of random ssDNA library and its primers
[0069] (a) Construct a random ssDNA library with a length of 79 bases:
[0070] 5'-TAGGGAATTC GTCGACGGAT CC-N35-CTGCAGGTCG ACGCATGCGC CG-3', wherein N represents any one of the bases A, T, C, G.
[0071] (b) Synthesize the forward primer:
[0072] Forward primer 1: 5′-TAGGGAATTC GTCGACGGAT-3′;
[0073] Forward primer 2: 5′-FAM-TAGGGAATTC GTCGACGGAT-3′;
[0074] (c) Synthetic reverse primer:
[0075] Reverse primer 1: 5'-CGGCGCATGC GTCGACCTG-3';
[0076] Reverse primer 2: 5'-biotin-CGGCGCATGCGTCGACCTG-3'.
Embodiment 2
[0077] Example 2: In vitro screening of nucleic acid aptamers
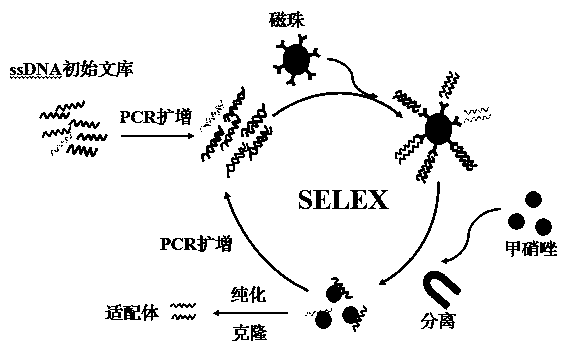
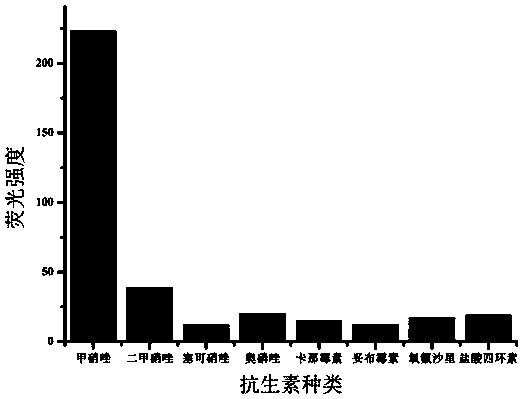
[0078] In order to screen out ssDNA aptamers with high affinity and specificity to metronidazole, a total of 10 rounds of nucleic acid aptamers were screened.
[0079] (a) The 25 μL PCR amplification system is shown in Table 1.
[0080] Table 1
[0081]
[0082] Amplification conditions: pre-denaturation at 95°C for 5 min; denaturation at 95°C for 30 s; annealing at 55°C for 30 s; extension at 72°C for 15 s; extension at 72°C for 5 min; 29 cycles.
[0083] (b) Main steps of in vitro screening: After washing the streptavidin-coupled magnetic beads with PBS buffer for 5 times, dissolve 200 μL of the initial library PCR product in 200 μL of binding buffer, and then add streptavidin The magnetic beads coupled with protein were gently shaken at room temperature for 2 h, then placed on a magnetic separator, the supernatant was removed, and the magnetic beads were washed 5 times with binding buffer, and then 300 mmo...
Embodiment 3
[0086] Example 3: Screened ssDNA clones and sequencing
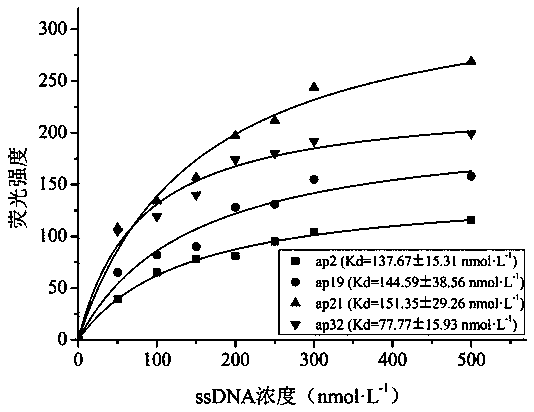
[0087] ssDNA clone sequencing: The ssDNA obtained after the final round of screening was amplified by PCR with forward primer 1 and reverse primer 1, and the entire amount of the amplified product was loaded on 3% agarose, and the PCR product was recovered. The purified PCR product was ligated with the pMD 18-T Vector vector according to the instructions of the T vector, and after overnight ligation at 16°C, it was transformed into Escherichia coli JM109 and cultured overnight. The correct transformants were verified by colony PCR and agarose gel, 39 positive clones were picked and their plasmids were extracted for sequence determination, and 39 aptamers with different sequences from ap1 to ap39 were obtained by sequencing.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com