Formula of probiotics for inhibiting pathogenic bacteria of colorectal cancer and screening method thereof
A technology for colorectal cancer and a screening method, applied in the field of biomedicine, can solve the problems of inability to use probiotics against colorectal cancer pathogenic bacteria, poor colonization ability, etc., so as to alleviate the serious threat to human health and the ecological environment , excellent performance, universal results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] S1. Constructed the human gut microbiome online warehouse (GMrepo, http: / / gmrepo.humangut.info / home) database, searched and downloaded the metagenomic data of normal people and colorectal cancer patients in the database, and obtained The double-end sequencing information (ENAaccession ID: ERP005534) of intestinal flora metagenomics (ENAaccession ID: ERP005534) of colon cancer patients (53 cases) and healthy people (61 cases), and then use Trimmomatic software to perform quality control on the data, remove low-quality sequences and adapters, and use Fastqc software evaluates the data after quality control, and obtains the metagenomic data after quality control;
[0039] S2. MetaPhIAn2 software was used for metagenome species annotation analysis, and the abundance information of intestinal flora species of normal people and colorectal cancer patients was obtained;
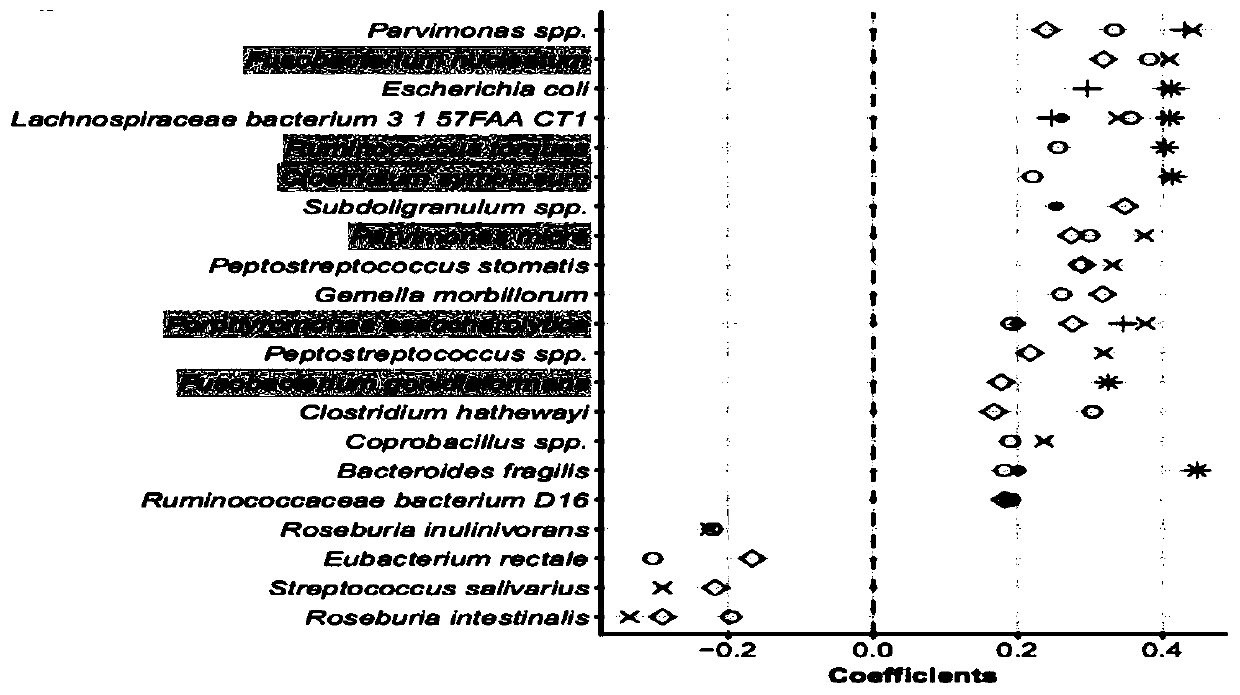
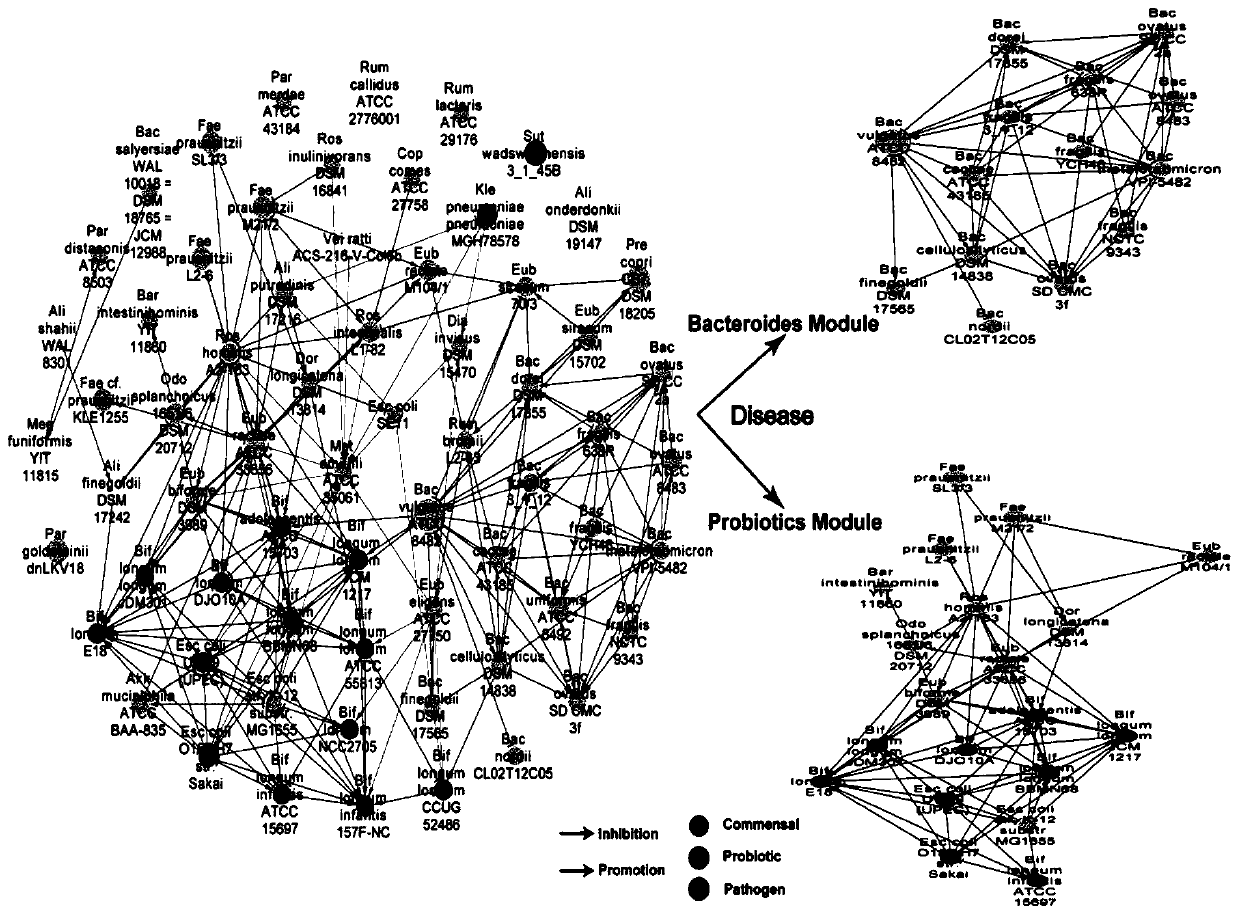
[0040] S3. Based on the metagenomic data after quality control in S1 and the abundance information in S2, L...
Embodiment 2
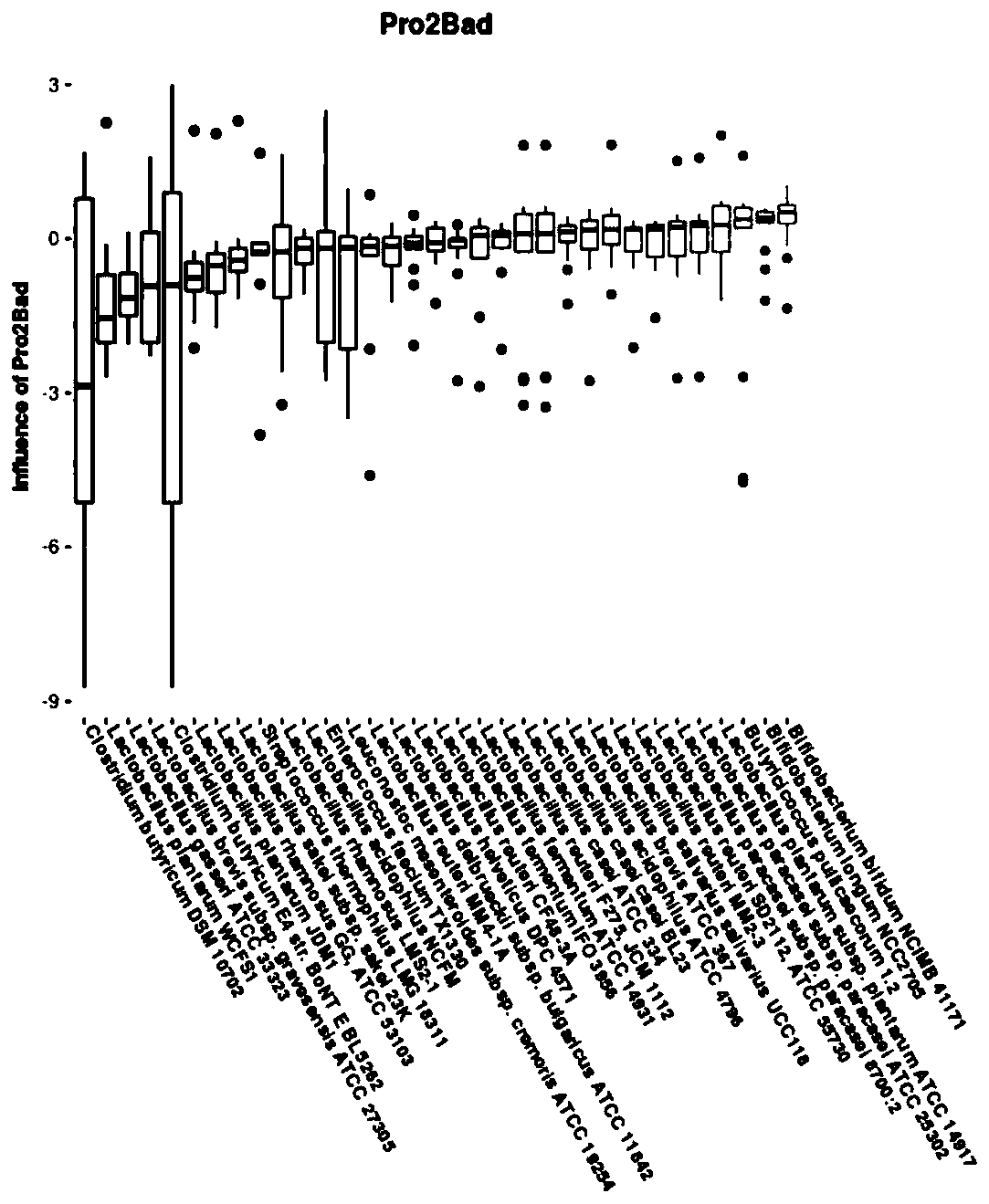
[0048] Compared with Example 1, the difference of this example is that the formula of probiotics in step S6 is: the content of Clostridium butyricum (Clostridium butyricum) is 1×10 9 CFU / ml, the content of Enterococcus faecium is 1×10 8 CFU / ml, the content of Lactobacillus brevis is 1×10 8 CFU / ml, the content of Lactobacillus plantarum is 1×10 8 CFU / ml, the content of Lactobacillus rhamnosus is 1×10 8 CFU / ml, the content of Lactobacillus sakei is 1×10 8 CFU / ml, the content of Leuconostoc mesenteroides is 1×10 8 CFU / ml. The end result is as Figure 6 shown, where Figure 6 -A is the abundance change of probiotics in this embodiment, Figure 6 -B is the abundance change of pathogenic bacteria in the present embodiment (wherein because the abundance of this pathogenic bacteria of Fusobacterium gonidiaformans is significantly higher than other pathogenic bacteria, so the abundance of Fusobacterium gonidiaformans corresponds to the ordinate on the right side of the figure, ...
Embodiment 3
[0050] Compared with Example 1, the difference of this example is that the formula of probiotics in step S6 is: the content of Clostridium butyricum (Clostridium butyricum) is 5×10 10 CFU / ml, the content of Enterococcus faecium is 5×10 9 CFU / ml, the content of Lactobacillus brevis is 5×10 9 CFU / ml, the content of Lactobacillus plantarum is 5×10 9 CFU / ml, the content of Lactobacillus rhamnosus is 5×10 9 CFU / ml, the content of Lactobacillus sakei is 5×10 9 CFU / ml, the content of Leuconostoc mesenteroides is 5×10 9 CFU / ml. The end result is as Figure 7 shown, where Figure 7 -A is the abundance change of probiotics in this embodiment, Figure 7 -B is the abundance change of pathogenic bacteria in the present embodiment (wherein because the abundance of this pathogenic bacteria of Fusobacterium gonidiaformans is significantly higher than other pathogenic bacteria, so the abundance of Fusobacterium gonidiaformans corresponds to the ordinate on the right side of the figure, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com