Upland cotton phosphatidylinositol specific phospholipase C gene GhPIPLC2-2 and application thereof
A technology of cotton phosphatidylinositol and phospholipase, applied in the field of plant genetic engineering, to achieve the effect of improving plant salt tolerance and salt resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1: high-fidelity PCR amplification GhPIPLC2-2 target gene
[0037] Take the young leaves and roots of 9807 four-leaf stage in upland cotton, refer to the instructions of the plant total RNA extraction kit (TIANGEN, Beijing, RN38), extract the total plant RNA, and use the PrimeScript RT Reagent Kit (TaKaRa, Dalian, RR047A) to invert The cDNA was obtained by reverse transcription with the recording kit. High-fidelity PCR amplification with specific primers.
[0038] The specific primer sequences are as follows:
[0039] GhPIPLC2-2-F: GGGGACAAGTTTGTACAAAAAAGCAGGCTTA ATGTCCAAACAAACTTACAGGG;
[0040] GhPIPLC2-2-R: GGGGACCACTTTGTACAAGAAAGCTGGGTT GATGAATTCAAAGTGCATAAGGAGC.
[0041] Underlines indicate BP-reactive homologous recombination sites.
[0042] The total volume of the PCR reaction is 50 μL, and the reaction system is: 10 μL 5×pfu Buffer, 1 μL dNTP, 1 μL forward primer, 1 μL reverse primer, 1 μL FastPfuDNA Polymerase, 1 μL cDNA template diluted ten t...
Embodiment 2
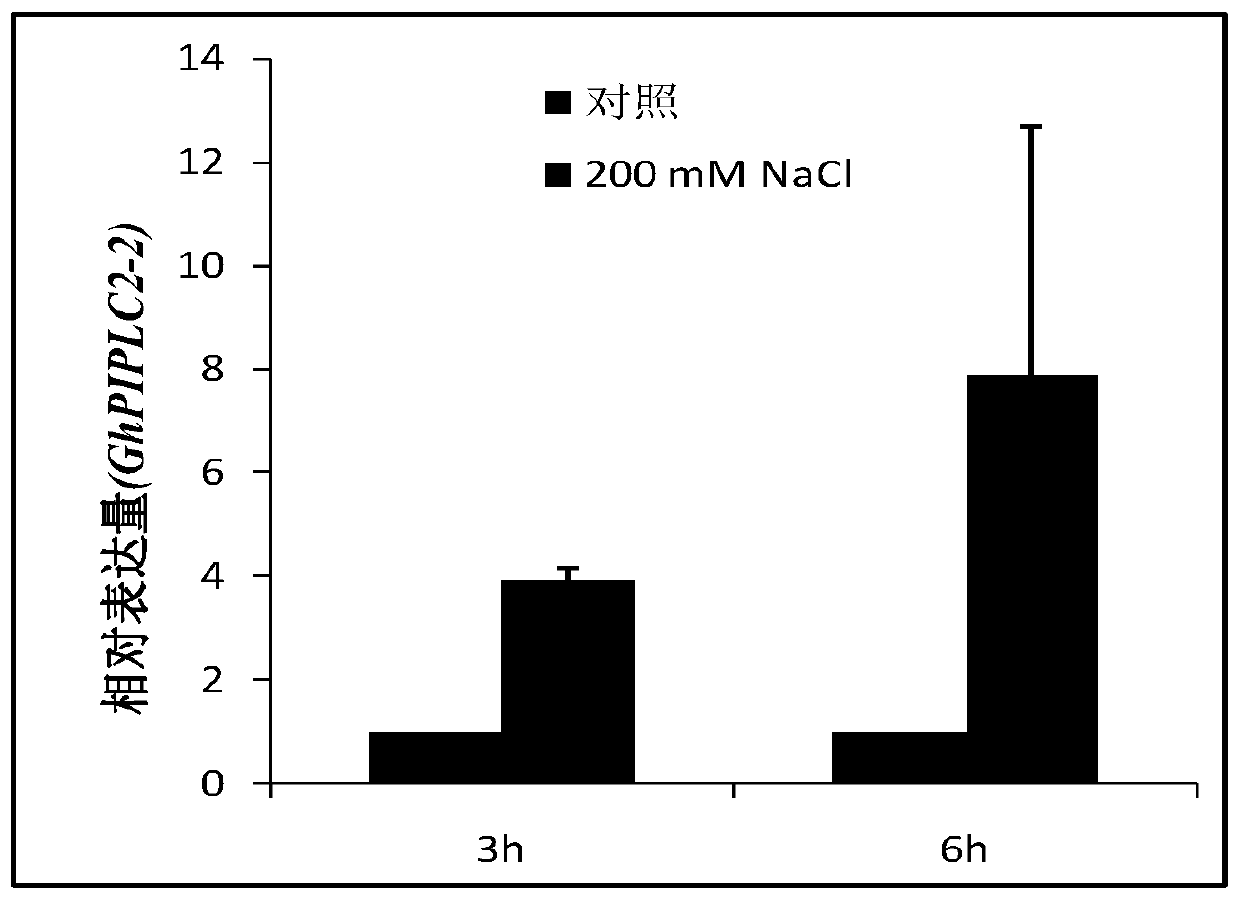
[0046] Example 2: Analysis of the expression pattern of the GhPIPLC2 gene under salt stress
[0047] The 9807 (Gossypium hirsutum L.Var. Zhong9807) seeds of upland cotton were sterilized and planted in the greenhouse under the following growth conditions: 30°C / 25°C, 60-70% relative humidity, 14 hours of light and 10 hours of darkness, and the photon flux density was 800 μmol m -2 the s -1 . When growing to the four-leaf stage, carry out salt (200mM NaCl) treatment, and the treatment time is 3h and 6h respectively. Total RNA was extracted and reversed into cDNA, diluted 10 times for real-time fluorescent quantitative PCR analysis. Upland cotton Histone gene (GenBankaccession number NC_006639) was used as the internal standard. Fluorescence quantitative PCR reaction kit is TAKARA's SYBR PremixEx TaqⅡ kit, the reaction is in 96System Fluorescence Quantitative Instrument, carried out 3 biological repetitions, the experimental results were measured by relative quantification ...
Embodiment 3
[0053] Embodiment 3: Verification of GhPIPLC2-2 gene function
[0054] 1. Construction of Arabidopsis expression vector
[0055] Take the young leaves and roots of 9807 four-leaf stage in upland cotton, refer to the instructions of the total plant RNA extraction kit (TIANGEN, Beijing, DP432), extract the total plant RNA, and use PrimeScript RT Reagent Kit (TIANGEN, Beijing, RN38) to invert The cDNA was obtained by reverse transcription with the recording kit. High-fidelity PCR amplification with specific primers. The specific primer sequences are as follows:
[0056] GhPIPLC2-2-F: GGGGACAAGTTTGTACAAAAAAGCAGGCTTA ATGTCCAAACAAACTTACAGGG;
[0057] GhPIPLC2-2-R: GGGGACCACTTTGTACAAGAAAGCTGGGTT GATGAATTCAAAGTGCATAAGGAGC (the underline indicates the BP homologous recombination site)
[0058] The total volume of the PCR reaction is 50 μL, and the reaction system is: 10 μL 5×pfu Buffer, 1 μL dNTP, 1 μL forward primer, 1 μL reverse primer, 1 μL FastPfuDNA Polymerase, 1 μL cDNA t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com